TRANSCRIPTION
Transcription is the process by which an RNA copy of a gene made from DNA or Transcription is the process by which stored information is taken from the genetic material and ultimately made available to the cell in the form of protein or RNA.

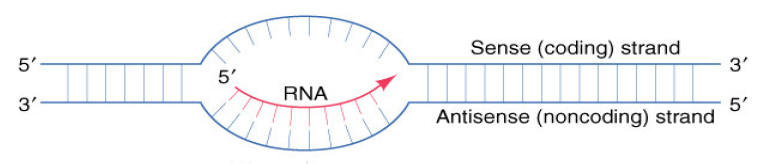
The DNA strand that is transcribed for a given mRNA is termed the template strand or Non-coding strand or Non-sense strand. Its complementary DNA strand will be referred to as the Non-template strand or coding strand or sense strand. The transcribed RNA has the sequence of non-template strand with the exception of U instead of T.
DNA DIRECTED RNA POLYMERASE:
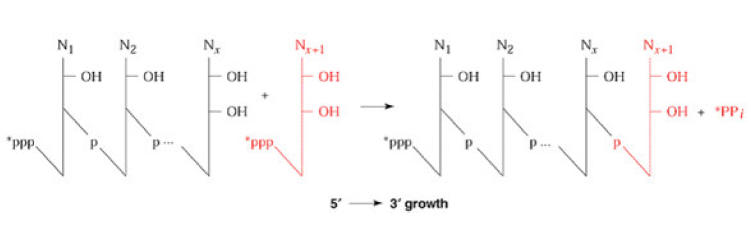
The enzyme DNA directed RNA polymerase begins the synthesis of RNA and adds ribonucleotides to the 3-end of RNA in which DNA molecules template strand determines the base sequence of RNA.
The ribonucleotide incorporation reaction requires energy provided by triphosphates of the four nucleotides. This reaction is driven by the hydrolysis of the pyrophosphate released upon incorporation of each nucleotide monophosphate into the growing RNA chain. The reactions are as follows:
(RNA)n + NTP -------------ΰ (RNA)n+1 + PPi
PPi --------ΰ 2Pi
Where
NTP ΰ Any triphosphates nucleotide
Pi ΰ Inorganic phosphate
PPi ΰ Pyrophosphate
n ΰ Number of nucleotide in RNA
The second reaction requires the enzyme inorganic pyrophosphatase to split the pyrophosphate, releasing the energy that drives the incorporation reaction. During RNA polymerase function two things occur simultaneously. First, DNA is being decoded. That is, the polymerase enzyme reads the deoxynucleotides of the anticoding or template, strand of DNA and base pairs those nucleotides with the appropriate ribonucleotides. Second, a phosphodiester bond is produced between the 3-position of the last ribonucleotide of the m-RNA and the 5-position of the new ribonucleotide to be incorporated. So, decoding of the gene and m-RNA synthesis occurs at the same time.
Mechanism:
Conditions for RNA Polymerase Activity (Pre-requisite):
The following are the requirements for DNA directed RNA polymerase functions
a) A template of a double stranded DNA
b) All four ribonucleotides
c) Mg2+ ions.
Unlike DNA polymerase, RNA Polymerase does not require primer.
Prokaryotic RNA Polymerase:
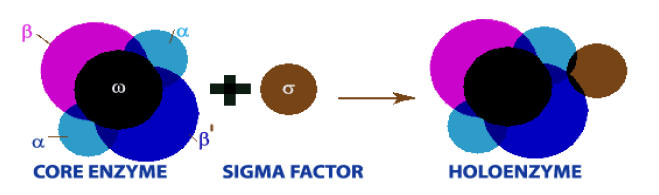
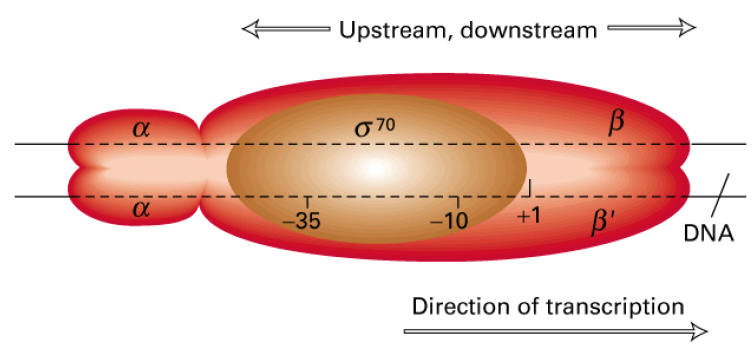
E.Coli RNA polymerase is a complex enzyme consisting of six subunits a2, b, b,w and s. The beta subunit (b) has a molecular weight of 150,000, Beta prime (b) 160,000, alpha (a) 40,000, omega (w) 11,000 and sigma (s) 70,000. The complex RNA polymerase enzyme of E.Coli, the holoenzyme, is composed of a core enzyme and a sigma factor. The core enzyme is composed of five subunits a2, b, b and w. Core enzyme can continue transcription after initiation but holoenzyme is necessary for correct initiation of transcription.
|
S.No |
Factors |
Functions |
|
1. |
s |
Recognition of Promoter sequence and aids the proper binding of RNA Polymerase to DNA initiation site. |
|
2. |
b |
Binding of RNA Polymerase to template |
|
3. |
b |
Polymerization function |
|
4. |
a, w |
Structural component of RNA Polymerase |
Eukaryotic DNA directed RNA polymerase:
The nuclei of eukaryotic cells contain three different RNA Polymerases, designated as I, II, and III. Each eukaryotic RNA Polymerases catalyzes transcription of genes encoding different classes of RNA. Subunit structure of yeast RNA polymerase is as follows:
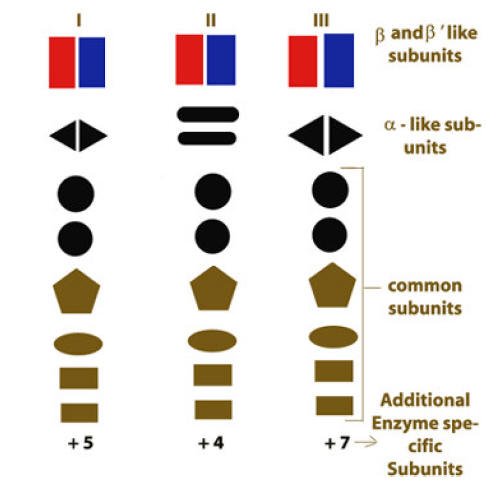
RNA Polymerase I :
RNA Pol- I is located in the nucleolus and is responsible for synthesis of precursor RNA (Pre-r-RNA), which is processed into the 28S, 5.8S and 18S r-RNAs. It accounts for nearly half the total RNA found in the cell. It is insensitive to a-amanitin
The above characteristic used to distinguish it from other polymerases. The enzyme is tightly regulated so that ribosome synthesis keeps pace with the cells protein requirements for growth, development and division. The complete enzyme includes two large polypeptide subunits and depending on the source, from 4 to 10 smaller subunits. Some of these smaller subunits are common to the other two polymerases. The Polymerases requires atleast two transcriptional factors for activity. These factors are needed for binding of the polymerase at the promoter site and to initiate transcription.
RNA Polymerase II:
The enzyme RNA Polymerase II produces all the pre-m-RNA of the cell and is thus responsible for the transcription of the largest part of the genome. RNA Pol-II also produces four small RNAs that take part in RNA Splicing [U1, U2, U3 and U4]. It present in nucleoplasm. It is very sensitive to mushroom poison a-amanitin.
It has been the most intensively studies of the three polymerases. The enzyme is composed of two large polypeptides and from 6 to 8 smaller polypeptides. This polymerase recognizes three different elements of a gene.
a) A selector sequence containing a TATA box and a short sequence.
b) An upstream promoter sequence
c) An enhancer sequence which may be located at different site in different genes.
Seven transcription factors required by RNA Pol-II for specific binding of the enzyme to the DNA promoter and to initiate transcription.
RNA Polymerase III :
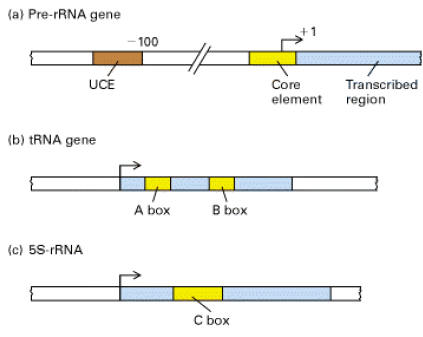
The enzyme RNA Pol-III present in nucleoplasm. It transcribes t-RNA genes, gene for 5S-r-RNA which is found in large ribosomal subunit (60S) of eukaryotes, genes whose RNA end products (e.g. Usn RNAs) assist in the processing of pre-RNAs by spliceosome and also genes of 7S RNA of the signal recognition particle (SRP) which is involved in the transport of proteins into the endoplasmic reticulum. It is the most structurally complex of the RNA polymerases. In yeast, the complete molecule is made up of 14 distinct polypeptides. Like other enzymes, it has two large polypeptides associated with smaller subunits. Few smaller subunits are common to the other polymerase. It is less sensitive to a-amanitin.
Template Independent RNA Polymerases :
There are few RNA polymerases are found in cells which does not require template for polymerization but they require pre-existing RNA contain unlike DNA directed RNA Polymerases.
Example:
a) t-RNA specific Nucleotidyl transferase:
It adds CCA sequence to 3-end of t-RNA during post transcriptional modification of pre-t-RNA.
b) Poly (A) Polymerase :
It adds poly A tail to 3-end of hn-RNA during post transcriptional modification of eukaryotic pre-m-RNA.
TRANSCRIPTION OF PROKARYOTIC RNA:
Transcription involves three stages:
1) Initiation
2) Elongation
3) Termination
1) Initiation:
Transcription begins when the DNA directed RNA Polymerase associates with the sigma factor to produce holoenzyme. The sigma factors allows the polymerase to bind specifically at the genes promoter sequence. There are two important promoter regions present. They are Pribnow box and -35 sequence.
Pribnow box (-10 region):
This is found 5 to 10 bases to the left or upstream from the first base copied into m-RNA. It contains a hexameric consensus sequence TATATT. Of which the T at 6th position called as conserved t because it is present in almost all prokaryotic promoters analyzed (96%).
-35 sequence:
It is present 16-19 bases upstream from the Pribnow box. It is a hexameric consensus sequence TTGACA. This region is the initial binding site for the s subunit of RNA polymerase.
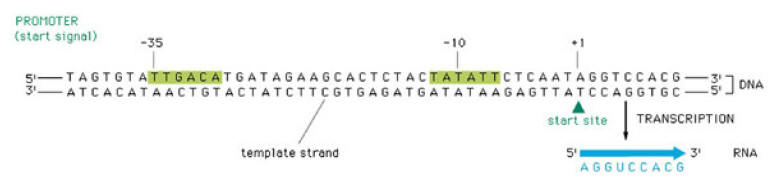
The first stage of transcription is the formation of an open promoter complex. First, the s subunit of the RNA polymerase recognizes and binds to -35 sequence followed by the binding of core enzyme.
Then the RNA Polymerase by virtue ofo its large size spans region of 17 to 19 base pairs and gets bound to the Pribnow box. This early stage structure of binding of RNA Polymerase (Holoenzyme) to the -10 region is called closed promoter complex.
RNA Polymerase inturn cause localized melting i.e. unwinding of DNA helix. This conformation stage is called open promoter complex. Unwinding spans in a region of about 10 basepairs from left end of the Pribnow box and extending about 20 base pairs after the position of the first transcribed base. The melting is necessary for the pairing of incoming ribonucleotides.
After open promoter complex formed, polymerization started. RNA Polymerase contains two nucleotide binding site called the initiation site and the elongation site [catalytic site]. The initiation site primarily binds purine trinucleotides (ATP or GTP). Therefore, the first nucleotide is to be incorporated is ATP or GTP. Thus, the first DNA base transcribed is either thymine or cytosine.
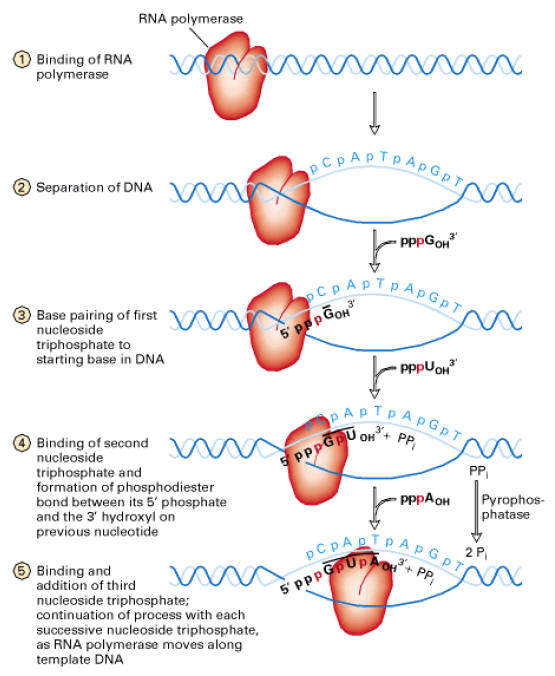
The initiating nucleoside triphosphates bind to the enzyme in the open promoter complex and form a hydrogen bond with a complementary DNA base. The elongation site is then filled with nucleoside triphosphates that are selected by its ability to hydrogen bond to the next base in the DNA strand. The two nucleotides are then joined together. The first nucleotide released from the initiation site. Then RNA Polymerase moves on DNA template exactly one nucleotide distance.
Once RNA Polymerase moves on, the DNA behind the enzyme closes as the hydrogen bonds of the DNA base pairs reforms. The enzyme reads the template strand in a 3 to 5 direction as it synthesizes m-RNA in a 5ΰ3 direction.
2) Elongation:
After several nucleotides (mostly eight) are added to the growing chain, RNA Polymerase undergoes a conformational change and the s subunit dissociates. Therefore, the chain elongation process is carried out by the core enzyme. Core enzyme continues reading the template strand and joining ribonucleotides by addition to the 3-end of the growing chain synthesis of m-RNA is therefore in the 5ΰ3 direction as the template is decoded in 3ΰ5 direction. The strands of double stranded nucleic acids, even in temporary hybrids such as DNA-RNA molecules, must be antiparallel if hydrogen bonding across the strand is to takes place.
The energy required for synthesis is provided by the triphosphates ribonucleosides. These ribonucleotides are the energy sources, building blocks and the information components for m-RNA synthesis.
3) Termination:
The last stage in m-RNA synthesis is chain growth termination. Synthesis of m-RNA is ended by any one of the following ways namely
a) Rho (r) independent termination
b) Rho (r) dependent termination
The DNA sequences, often referred to as transcription terminators are either rho-dependent or rho-independednt. In either case, a so called stem-loop or hairpin structure, is formed. RNA synthesis terminates shortly after this structure is formed.
The stem-loop forms at the 3-end of the m-RNA, because at the 5-end of the template DNA an unusual sequence of nucleotides occurs. This sequence is known as dyad symmetry i.e. inverted base sequence with central repeat sequence. That is, read in a 5ΰ3 direction, the DNA nucleotide sequences of the two strands are identical. For example,
5 GGCTCCTTTTGGAGCC 3
3 CCGAGGAAACCTCGG 5
When m-RNA is transcribed from the template strand (3ΰ5), the resulting sequence is
5 GGCUCCUUUUGGAGCC 3
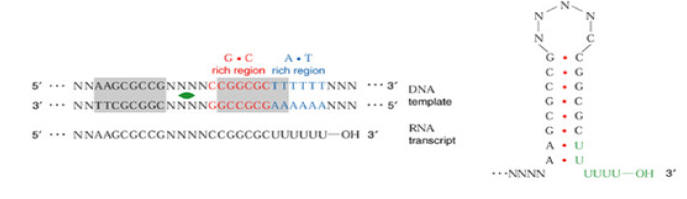
The molecule is self complementary, so a hairpin or stem loop can form.
A) Rho-independent Termination:
In Rho-independent termination, the template inverted repeat of DNA is followed by a series of adenines. This series produces a run of perhaps half a dozen Uracils in the m-RNA. So, the m-RNA in rho-independent termination has the following structure.
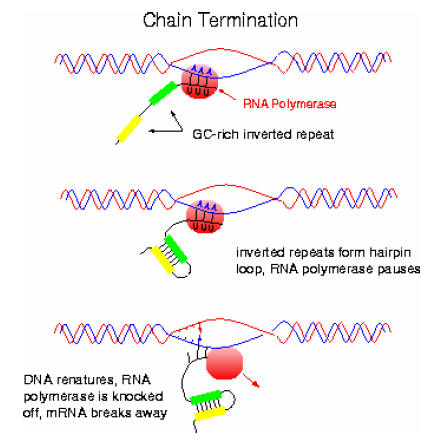
At the point where the poly U sequence is attached to the DNA sequence, the hybrid DNA-RNA is unusually weak (A-U bonds are weak) and it requires very little energy to break the hydrogen bonds holding the two strands together. When separation occurs, m-RNA synthesis, transcription stops. This type of termination is rho-independent; no termination factor is required.
B) Rho dependent Termination:
Rho dependent termination also uses a hairpin m-RNA formation but dissociation of the DNA-RNA hybrid needs the assistance of the protein rho and no poly U follows the hairpin. Rho, a tetramer of about 5 kdaltons binds to RNA Polymerase and brings about the excision of RNA transcript by a mechanism which is not fully understood. It is proposed that first rho protein binds to the 5-end of a nascent RNA chain and then moves along the RNA, using the hydrolysis of ATPs to provide the necessary energy. Then, when RNA Polymerase pauses at certain sites having high GC sequences or stem-loop structure it catches up and binds with RNA Polymerase. When rho binds with RNA Polymerase it assume ATPase and brings about the hydrolysis of RNA chain after which the rho factor and the enzyme dissociates from the template.
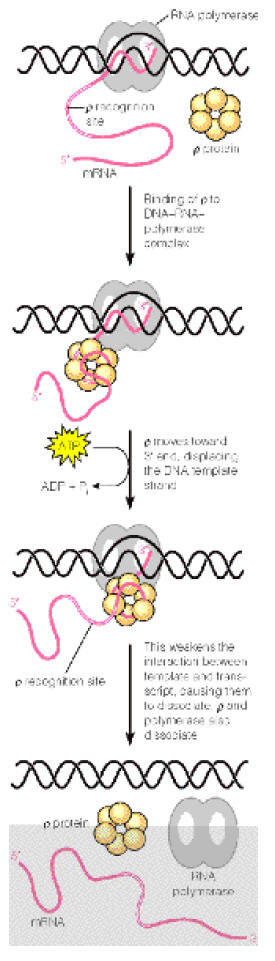
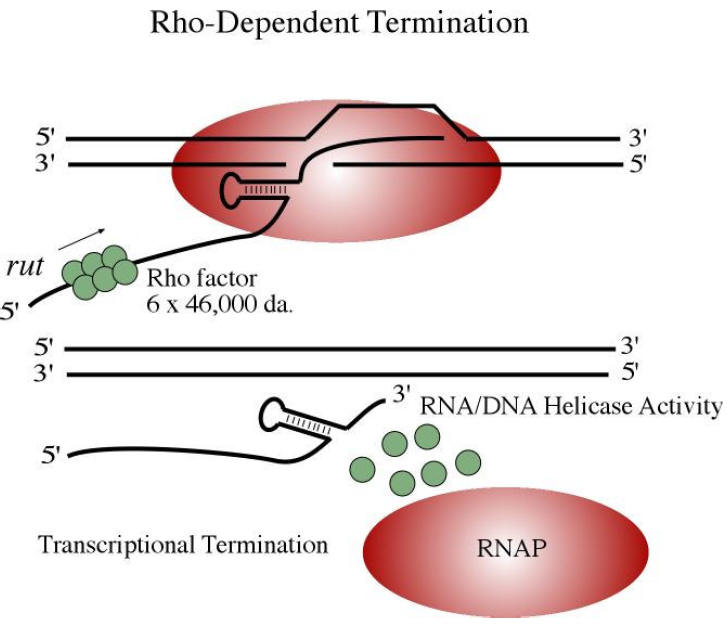
It also appears that termination is not absolutely rho dependent or rho-independent. Rather, rho-independent termination can utilize rho and rho dependent termination can proceed in the absence of the protein.
Regulation of Prokaryotic Transcription:
(Control of Prokaryotic Transcription)
There are different ways in which transcription of Prokaryotes controlled namely
a) Promoters activity
b) Repressors activity
c) Catabolite Repression
d) Dual Positive and Negative control
e) Attenuation
f) Stringent Response
a) Promoters Activity:
There are different types of promoter sequences present in DNA molecules. Depending upon the conservity of the sequences match with Pribnow box, promoters classified into two different types. They are strong promoters and weak promoters. Strong Promoters have the sequence which resemble almost similar to Pribnow box whereas weak promoters sequence varied to larger extent. There are different types of s - molecules which intern has different extent of affinity to promoter sequences. So strong promoters sequence with high affinity s - molecules will provide increased transcription rate and vice versa.
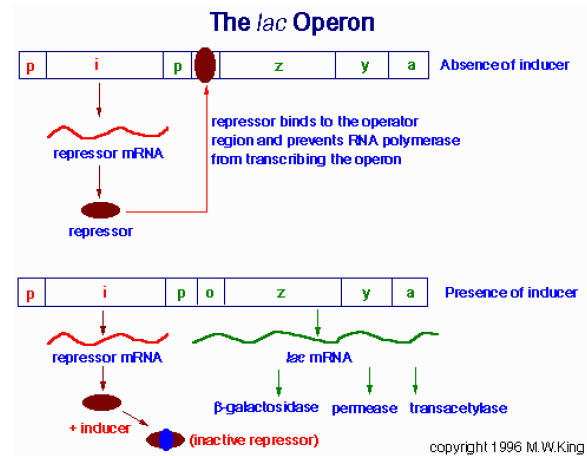
b) Repressor Activity:
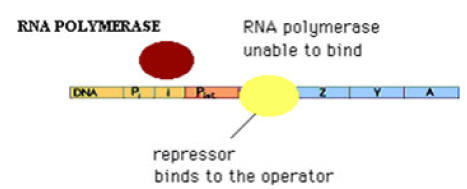
Repressors will bind to the operator region. So, that RNA Polymerase is unable to bind to the promoter region because promoter and operators regions are overlapping to each other. Because of this, there is no expression of genes example Lac Operon. At low lactose concentration, repressors prevent the expression of Lac Operon.
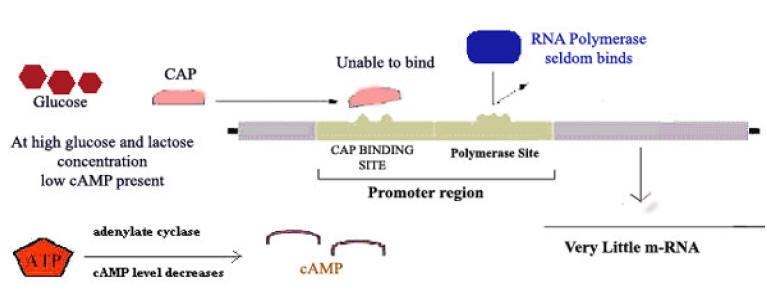
c) Catabolite Repression:
Glucose is E.Colis metabolite of choice; the availability of adequate amounts of glucose prevents the full expression of genes specifying proteins involved in the fermentation of numerous other catabolites including lactose, arabinose and galactose, even when they are present in high concentrations. This phenomenon, which is known as catabolite repression, prevents the wasteful duplication of energy producing enzyme systems.
Ex: Lac Operon
When glucose concentration increased inside cell then concentration of cAMP decreased which intern decrease cAMP- CAP complex (Catabolic gene Activator Protein). This intern decreases the rate of transcription. Thus catabolite represses the gene expression
d) Dual Positive and Negative Control:
Example: Ara Operon
Ara C protein plays dual positive and negative control on Ara operon.
In the presence of arabinose, the Ara C protein binds to the ara I region and when bound to cAMP, the CAP protein binds to a site adjacent to ara I. This binding stimulates the transcription of the structural genes.
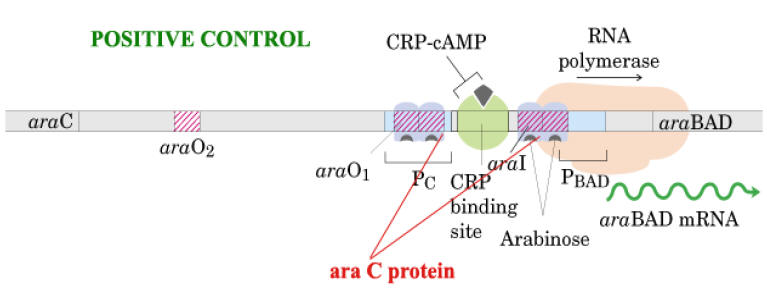
In the absence of arabinose, the Ara C protein binds to both ara I and ara O regions, forming a DNA loop. This binding prevents transcription of the ara operon.
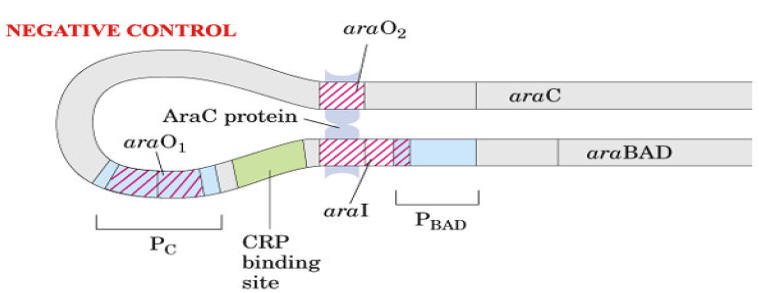
e) Attenuation:
Attenuation is the process for regulation of prokaryotic gene expression. It occurs in anabolic in anabolic operons i.e. the gene products are responsible for the synthesis of some compounds mainly aminoacids like tryptophan, Histidine, Leucine etc., these operons have a sequence known as Leader sequence inbetween operator and structural genes. This sequence contains four segments. These segments have the sequence such that they can form stem and loop structure. When 3rd and 4th segments forms stem and loop structure, termination of transcription would occur. But when 2nd and 3rd segment forms stem and loop structure, no transcriptional termination and transcription continues.
In prokaryotes, translation is closely coupled with transcription i.e. during transcription after small segment of gene transcribed; ribosome attached to it and initiates translation.
Example: Trp Operon
Segment 1 of transcribed m-RNA of Trp Operon contain codons for Tryptophan which intern regulate transcription. When tryptophan is abundant, segment 1 of the trp m-RNA is fully translated. Segment 2 enters the ribosome which enables Segment 3 and 4 to base pair. This base paired region signals RNA Polymerase to terminate transcription.
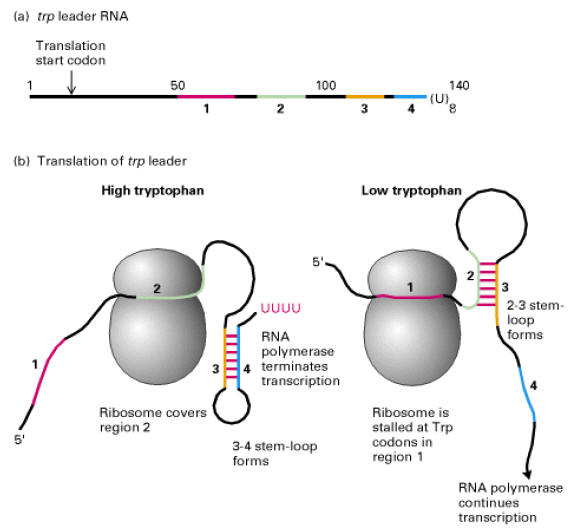
When tryptophan is scarce, the ribosome stalled at the codon of segment 1. Segment 2 interacts with segment 3 instead of being drawn into the ribosome and so segments 3 and 4 cannot pair. Consequently, transcription continues.
Thus the 2-3 segments stem and loop structure known as attenuator. This process is known as attenuation.
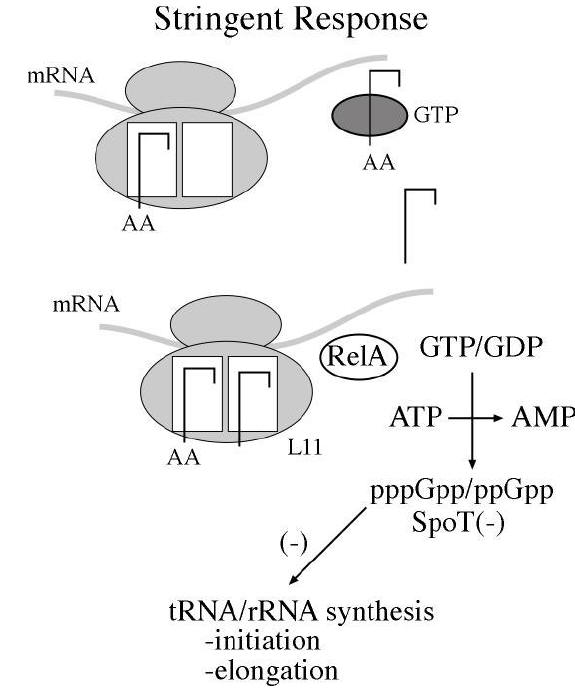
f) Stringent Response:
Stringent Response controls transcription of r-RNA and t-RNA. Guanosine tetra phosphate [ppGpp] is the prime component for stringent response. When charged t-RNA not available for codons, uncharged t-RNA attached to A Site of ribosome. So that ribosome stalls at that site. This favors synthesis of ppGpp by rel A. ppGpp then binds with RNA Polymerase and inhibit its action towards promoters for r-RNA and t-RNA genes. But transcription of genes for amino acid synthesis, lac operon and ara operon are initiated.
When charged t-RNA [aminoacyl t-RNA] available, then the level of ppGpp decreased by the action of spot. Once ppGpp concentration decreased, inhibition of RNA Polymerase released and thus transcription of r-RNA and t-RNA occurs.
EUKARYOTICE TRANSCRIPTION:
In eukaryotes, different RNA Polymerases involved in transcription of different type of RNAs. Hence mechanism differs. Transcription by each RNA Polymerases studied separately. They are
a) Transcription by RNA Pol I
b) Transcription by RNA Pol II
c) Transcription by RNA Pol III
A) TRANSCRIPTION BY RNA POLYMERASE - I:
RNA Pol I dedicated to the synthesis of only one type of RNA molecule, called pre-r-RNA. The primary pre-r-RNA transcript is processed into the 18S, 5.8S, and 28S rRNAs found in vertebrate ribosomes or their functional equivalents in other eukaryotes.
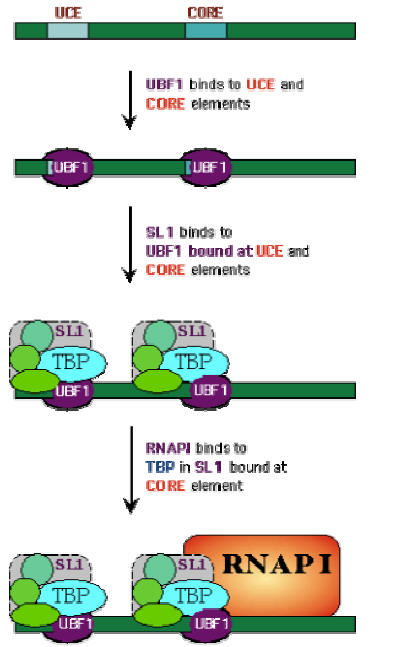
The control region of pre-RNA transcription units contains a core promoter element which overlaps the start site and an upstream control element (UCE) located ~100 base pairs upstream. Upstream binding factor (UBF) binds to the UCE and core element and the two bound molecules are through to make protein-protein interactions causing the intervening DNA to loop out. Selectivity factor (SL1) then binds to the UBF DNA complex and the remaining free segment of the core element. SL1 is a multimeric protein composed of TBP and three TBP-associated factors with Mws of 110, 63 and 48kDa. It is also species specific factor. Finally, RNA Polymerase I binds, completing assembly of the initiation complex.
After the formation of initiation complex, Pol-I unwind DNA and initiates transcription. Transcription is then elongated with RNA Pol I alone. Finally, transcription is terminated in a rho dependent termination manner.
B) TRANSCRIPTION BY RNA POLYMERASE II :
Promoters for RNA Pol II
1) TATA box [Goldberg Hogness box] [-25 region]:
It is located about 25 base pairs upstream from the transcription start site and has a consensus sequence TATAAAT. The TATA box is usually flanked by high GC sequences.
2) GC box:
It is located about 40 basepairs upstream from transcription start site. It has consensus sequence GGGCGG.
3) CAAT box:
It is located about 75 base pairs upstream from the transcription start site and has a consensus sequence GGT/CCAATCT.
4) Upstream Activating Sequence:
These sequence are also known as hypersensitive sites and are thought to influence many m-RNA synthesis for many specific proteins.

Transcription by RNA Pol II occurs in four stages. They are
I Formation of Initiation Complex
II Initiation
III Elongation
IV Termination
I) Formation of Initiation Complex:
Initiation complex begins with the binding of transcription factor TF II D to the TATA box. TF II D is composed of one TATA box binding subunit called TBP and more than either other subunits (TAFs), represented by one large symbol. TF II A binds to TF II D promoter complex to form DA complex. TF II B then binds to D A complex, followed by binding of a preformed complex between TF II F and RNA Polymerase II. Finally, TF II E, TF II H and TF II J must add to the complex, in that order, for transcription to be initiated. Initiation by RNA Pol II requires hydrolysis of the b - g bonds of ATPs. One of the last factors to add to the complex, TF II H, which has DNA helicase activity, can use the energy from hydrolysis of ATP to separate the strands of the duplex template DNA. This protein is suspected to mediate unwinding of the strands at the start site allowing the Polymerase to initiate transcription. TF II H also has a protein kinase activity, which can transfer the g - phosphates of ATPs to multiple serines in C-terminal repeat domain (CTD) of the largest RNA Pol II subunit.
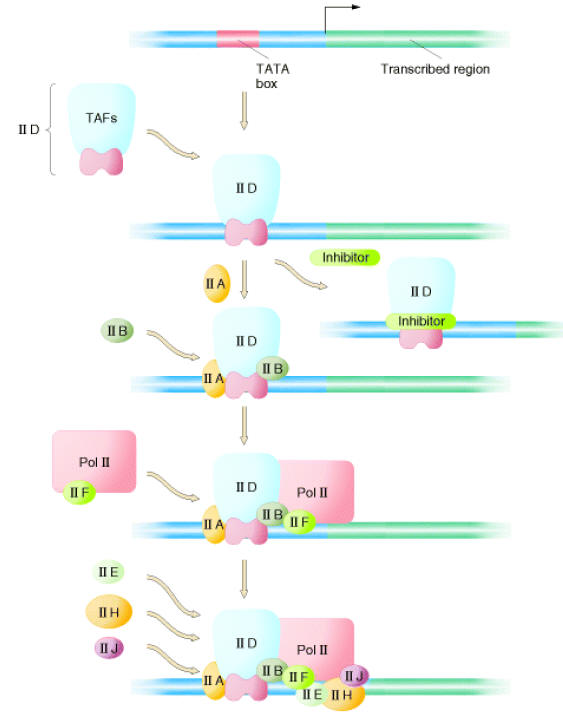
II ) Initiation:
After open promoter complex formation in which the template strand is exposed for transcription, initiation occurs. RNA Pol II contain two nucleotide binding sites, called the initiation site and elongation site. The initiating nucleoside triphosphates bind to the enzyme and forms hydrogen bond with a complementary base in DNA at initiation site. Elongation site is then filled with nucleoside triphosphates that are selected by its ability to hydrogen bonded to the next base in the DNA strand. The two nucleotides are then joined together through phosphodiester bond and the first base is released from the initiation site. Then RNA Polymerase moves in the relative direction so that enzyme shifted exactly by one nucleotide distance. After initiation TF II E released from initiation complex.
III) Elongation:
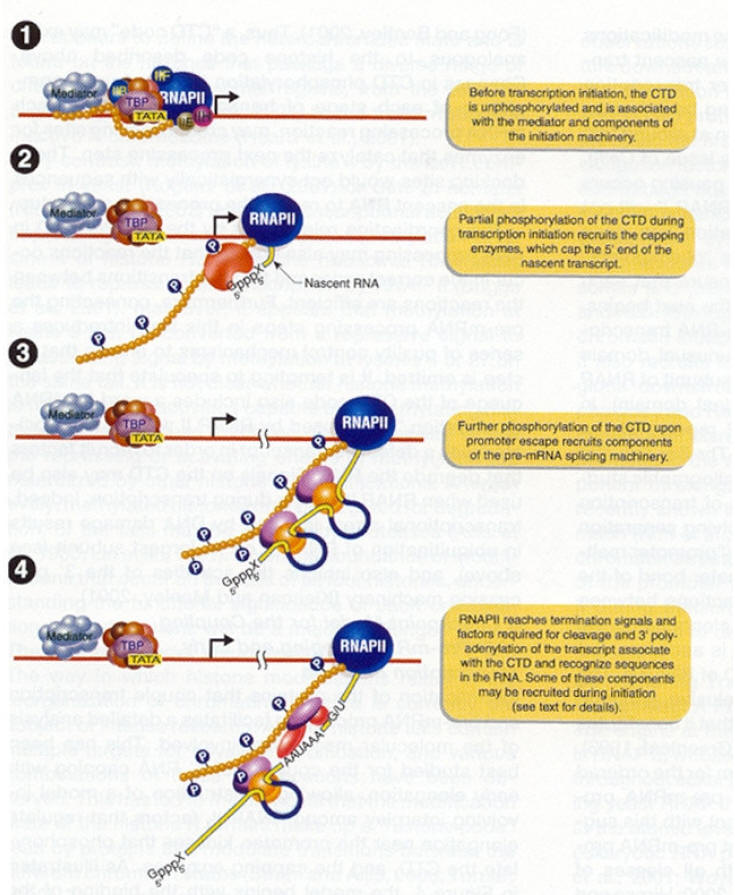
After few nucleotides added to the growing pre-mRNA chain, TF II H adds phosphate to serine residues in CTP of Pol II. This phosphorylation releases attachment of CTD to TF IID. After phosphorylation, RNA Polymerase II can move freely on DNA template and TF II H dissociates. Therefore, the chain elongation process is carried out by the RNA Pol-II and TF II J and F. RNA Pol-II unwinds DNA continuously as the enzyme extends the growing RNA chain. Nascent RNA chain grew in 5ΰ3 direction.
IV) Termination:
Termination does not require rho factor. Termination is rather carried out by the core enzyme itself by virtue of its ability to recognize certain sites in the DNA template which are called termination signal sites. This has three distinguishing features. They being
a) It has a inverted repeat base sequence containing central repeat sequence which allows the formation of stem and loop configuration leading to the excision of the RNA transcript.
b) It has high GC regions
c) It has high AT regions
All the above mentioned structural features of the termination site helps in the formation of hairpin structure in growing RNA chain which gets easily dissociated from DNA template and hence its termination.
C) TRANSCRIPTION BY RNA POL III:
i) Transcription of t-RNA genes:
First, TF II C, a large multisubunit protein, binds with high affinity to the B box promoter and with low affinity to the A box. TF III C acts as an assembly factor for binding the trimeric TF III B to any DNA sequence upstream of the t-RNA gene. TF III B is made up of three subunits. One is TBP. The second called BRF (TF II B related factor) is similar in sequence to TF II B. The third subunit of TF III B is a 90 kD polypeptide called B. Once TF III B binds, then RNA Polymerase can bind and TF III C is released. RNA Polymerase then initiates transcription in the presence of ribonucleoside triphosphates. Enzyme does not require hydrolysis of an ATP b - g bond similar to RNA Pol I. After initiation, RNA Polymerase elongates transcription and finally terminates transcription in a rho independent termination mechanism.
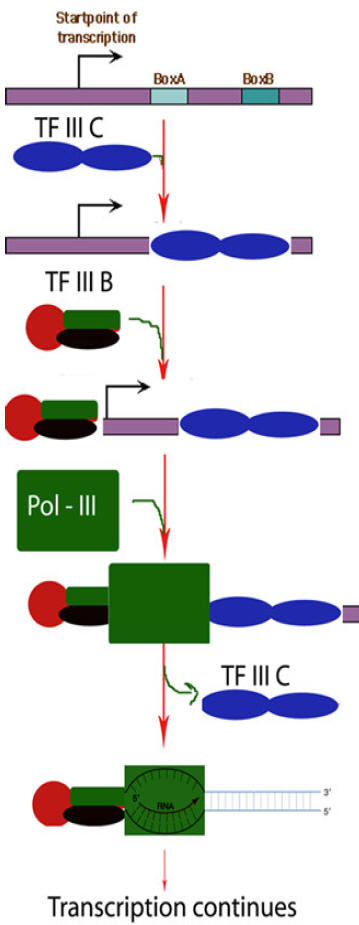
ii) Transcription of 5S r-RNA gene:
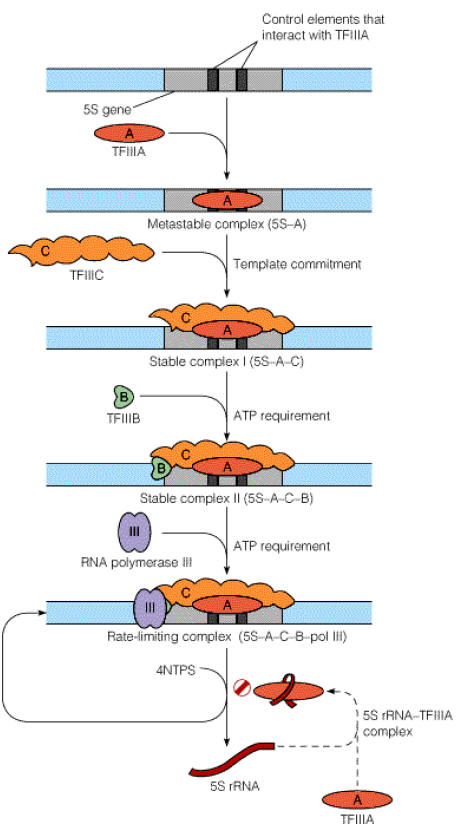
Synthesis of 5S r-RNA is initiated by binding of TF III A to the C box. Once TF III A has bound, TF III C binds to the gene at a similar position relative to the start site as when TF III C binds to a t-RNA gene. TF III B then binds, interacting analogously with TF III C as it does in a t-RNA gene. Once TF III B has bound, RNA pool III and binds and initiates transcription. TF III A thus acts an assembly factor for binding of TF III C; TF III C then act as an assembly factor for TF III B. RNA Pol III elongates transcription and terminate it in a rho dependent termination mechanism.
REGULATION OF EUKARYOTIC TRANSCRIPTION:
Eukaryotic transcription regulated with the help of any one of the
following components, namely,
i) Cis-acting elements
ii) Trans-acting elements (Transcriptional factors)
iii) Hormones
iv) Antiterminants
i) Cis-acting elements:
Cis-acting elements are specific sequence in DNA. They are of different types like promoter, proximal promoter element, enhancer and silencers. When specific components bind to promoter, promoter proximal element and enhancer, transcription initiated and rate increased whereas when it bound to silencer, transcription suppressed.
ii) Trans-acting elements:
Transcriptional factors like TF IIB, SL1, TFIIIB etc., are transacting elements. When transcriptional factors are present along with polymerase then transcription occurs. When their concentration decreased, transcriptional rate also decreased.
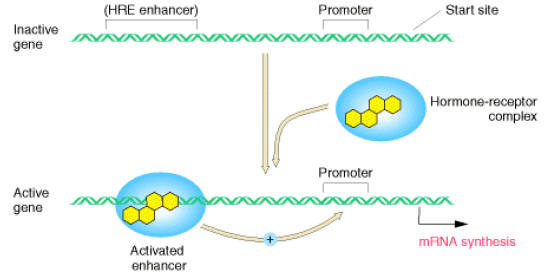
iii) Hormones Steroid Hormones:
Steroid Hormones enters into the cell and it binds with receptor to form hormone receptor complex. Then HR complex translocates inside the nucleus. Within the nucleus, HR binds with hormone response element which intern stimulates transcription. Absence of these hormones decrease transcription rate.
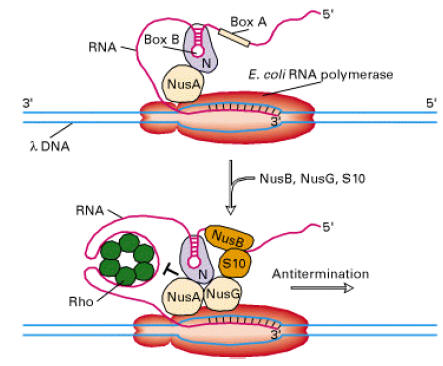
iv) Antiterminators:
In some genes, termination signal sequence present even after promoter itself. Depending upon the need of specific gene products, this termination signal may cause pretermination. In some cases, this premature termination was prevented by certain factors. They are referred as Antiterminators.
E.g Expression of C-myc gene
Presence of growth factors includes expression of C-myc gene which intern prevents the premature termination. So growth factor act as Antiterminators.
POST TRANSCRIPTIONAL MODIFICATION OR PROCESSING:
The immediate products of transcription, the primary transcripts, are not necessarily functional entities. In order to acquire biological activity, many of them must be altered in several ways.
i) by the exo and endo nucleolytic removal of polynucleotide segments
ii) by appending nucleotide sequence to their 3 end and 5-ends and
iii) by the modification of specific nucleosides
The three major classes of RNAs mRNA, rRNA and tRNA, are altered in different ways ini prokaryotes and in eukaryotes. These modifications after transcription are referred as Post transcriptional modifications or processing.
m-RNA PROCESSING:
Prokaryotic m-RNA:
In prokaryotes, most primary m-RNA transcript functions in translation without further modification. Ribosomes in prokaryotes usually commence translation on nascent m-RNAs itself. So, prokaryotic m-RNA does not undergo post transcriptional processing.
Eukaryotic m-RNA processing:
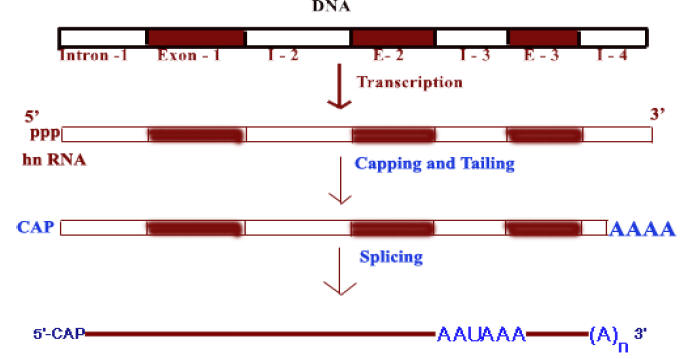
In eukaryotes m-RNAs are synthesized in nucleus while translation occurs in the cytosol. Apart from spatial segregation there also exists a finite temporal lag i.e. to say the transcription and translation does not go hand by hand. Infact, m-RNA synthesized as heterogeneous nuclear RNA [hnRNA] or pre-m-RNA which is the primary m-RNA transcript in the nucleus. It thereafter undergoes extensive post transcriptional processing while still in the nucleus, to form mature m-RNA which then gets transported to cytosol to get associated with ribosomes for the translation process to commence. Processing of m-RNA involves the following stages,
a) Capping
b) Tailing
c) Splicing
d) Methylation
a) Capping:
All eukaryotic m-RNAs have a cap structure at the 5end consisting of a 7-methyl guanosine residue join to the transcript via 5-5 triphosphates bridge. The cap structure is attached to the 5-end of the growing transcript by guanylyl transferase before it is greater than 20 nucleotides long. There are three types of capping
Cap O:
When the two leading nucleosides are not methylated at 2 position, are called cap O type which occurs predominantly in unicellular eukaryotes.
Cap 1:
When the first nucleoside following the 7-mehtyl guanosine methylated at the 2position, it is called cap 1 structure. Such capping occurs in most of the multicellular organisms.
CAP 2:
When the first two nucleosides following 7-methyl guanosine is methylated at 2 position, it is called cap2 structure and it is found in some eukaryotes.
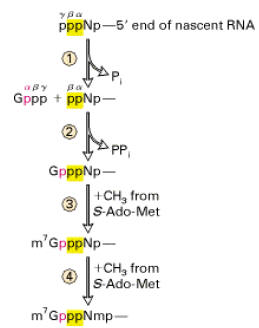
Significance of capping:
I) Enhancement of translation ability of m-RNA. Recent studies showed that capping of m-RNA is essential for binding to the smaller subunit of ribosomes.
II) Capping protects m-RNA from ribonuclease (RNase)
CAP STRUCTURE:
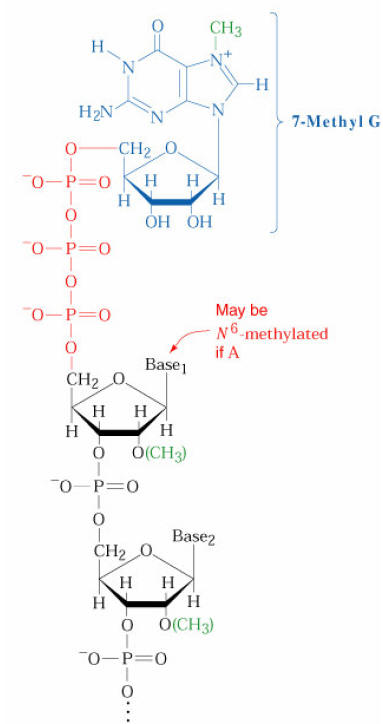
b) Tailing:
Tailing is a process in which poly A tail with around 200 adenosine residues attached to 3-end of hnRNA. Events in Tailing shown in the following diagram.
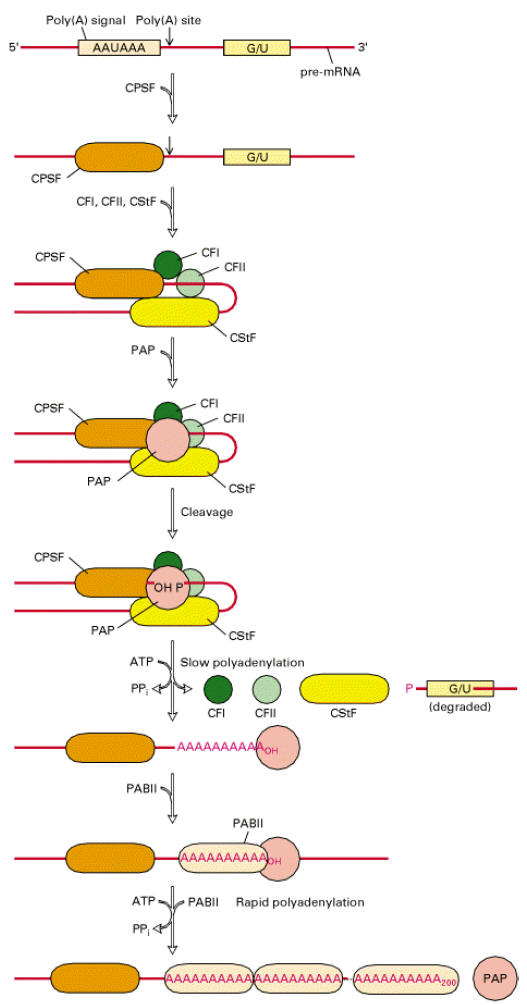
Significance:
Experimental evidences are shown that poly A stabilizes m-RNA. m-RNAs which have poly A tail have greater life time in cytosol whereas other m-RNAs which have no poly A tail have lifetime less than 30 minutes in cytosol.
c) Splicing:
The most striking differences between eukaryotic and prokaryotic structural genes are that the coding sequences of most eukaryotic genes are interspersed with unexpressed regions. Because of this eukaryotic genes known as split genes.
Splicing reaction involves the removal of nonfunctional or non-coding introns and joining of functional or coding exons.
Exons: They are the coding or functional sequences (or) expressed sequences of gene which gets transcribed in the primary RNA transcript and is retained in the final mature m-RNA.
Introns: They are the noncoding or nonfunctional intervening sequences (IVs) of gene which gets transcribed in the primary RNA transcript but are not retained in the mature m-RNA as a result of splicing reactions.
Mechanism of splicing:
For splicing to occur the following sequences are necessary at the splice site junctions and in the introns. At the splice junction AAGU is the highly conserved sequence at the 5 boundary and AGG at the 3 boundary. In the introns, a conserved sequence of CURAY has been found about 20 to 50 residues upstream at the 3 splice site.
Steps:
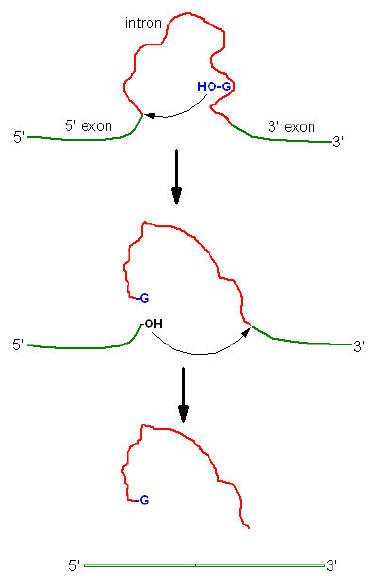
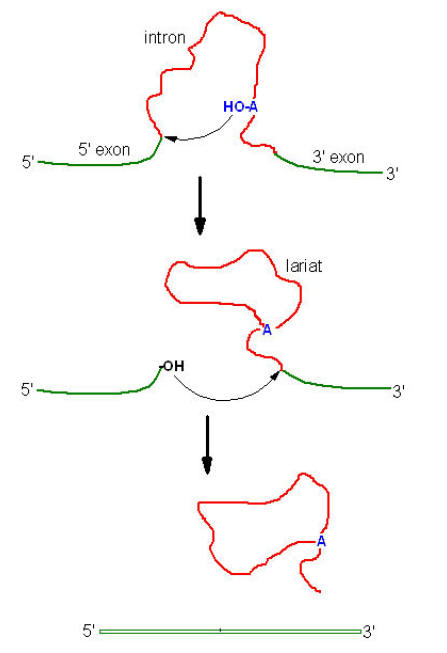
i) First a 2-5 phosphodiester bond is formed between an introns adenosine residue and its Guanosines 5-terminal phosphate group with the concomitant release of the 5-exons. The introns thereby assume Lariat structure.
ii) The adenosine residue at the Lariat branch has been identified as the A in the CURAY sequence [where R represent purines and Y represent pyrimidines] which is highly conserved in vertebrate m-RNA.
iii) Now free 3-OH group of the 5-exon forms a phosphodiester bond with the 5-terminal phosphate of the 3-exon, yielding the splice product. The introns is eliminated in its Lariat form.
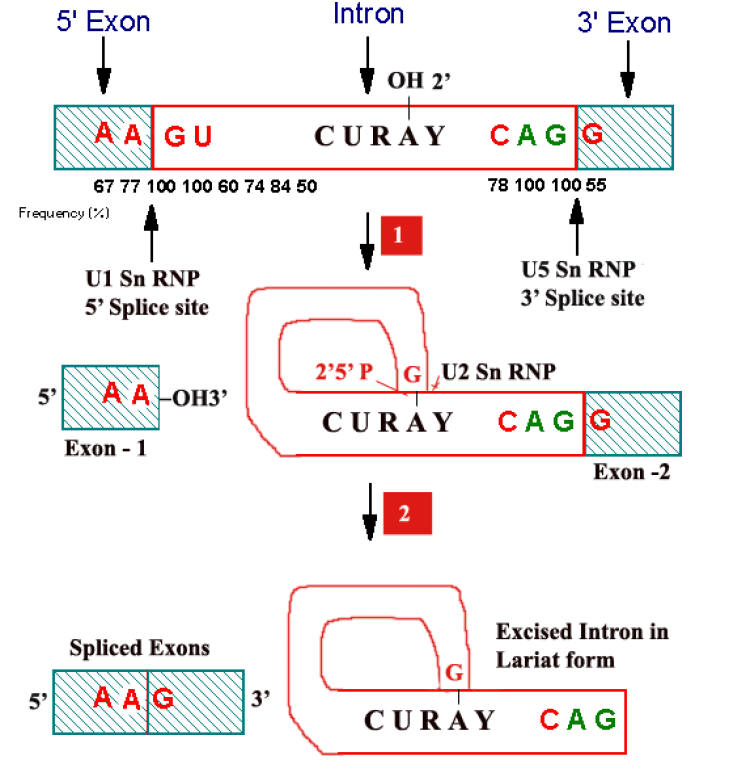
Role of SnRNPs:
Splicing reactions are mediated by small nuclear ribonucleoproteins [SnRNPs; pronounced as snurps] which are a complex of SnRNAs and proteins. There are more than 8 types of SnRNPs. Of which U1SnRNP, U2SnRNP, U5SnRNP, U4 and U6 SnRNPs are best characterized. U1SnRNP recognizes 5 splice junction, U2SnRNP then recognizes the introns region that forms the branch point while U5SnRNP recognizes the 3 splice junction.
SPLICEOSOME:
The large RNA-protein body, within which, nuclear m-RNA precursor is processed to remove introns. It is of 50S-60S particle. Spliceosome brings together a pre-m-RNA, the foregoing SnRNPs and a variety of pre-m-RNA binding proteins. Note that the spliceosome, which consists of 5 RNAs and atleast 50 polypeptides, is comparable in size and complexity to the E.Colis large ribosomal subunit 50S. Spliceosome carry out the splicing reactions of pre-m-RNA.
iv) Methylation:
During or shortly after the synthesis of vertebrate pre-m-RNAs, approximately 0.1% of their A residues are methylated at N6. These m6As tend to occur in the sequence RRm6ACX, where X is rarely G. Although the functional significance of these methylated As is unknown, it should be noted that a large fraction of them are components of the corresponding mature m-RNAs.
RNA EDITING:
RNA editing is a process in which the sequence of a pre-m-RNA altered. As a result, the sequence of the corresponding mature m-RNA differs from the exons encoding it in genomic DNA.
Certain m-RNAs from a variety of eukaryotic organisms have been found to differ from their corresponding genes in several unexpected ways, including CΰU and UΰC changes, the insertion or deletion of U residues and the insertion of multiple G or C residues.
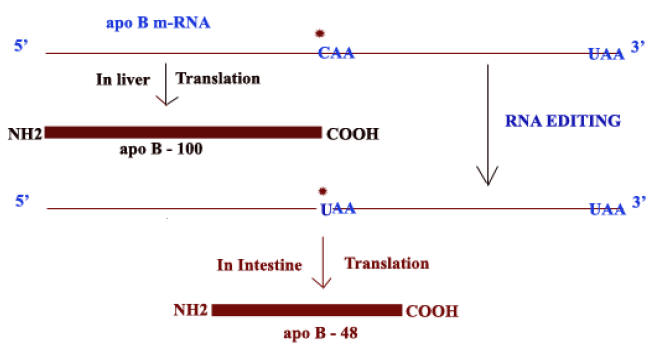
RNA editing of apo-B m-RNA:
The apo-B m-RNA produced in the liver has the same sequences as the exons in the primary transcript. This m-RNA is translated into Apo B-100 whereas in the apo-B m-RNA produced in the intestine, the CAA codon in exon 26 is edited to a UAA stop codon. As a result, intestine cells produce Apo-B-48. Production of different apo-B from the same gene is because of RNA editing. Addition of U residues to m-RNA achieved with the help of guide RNAs (gRNA) during RNA editing.
Example: m-RNA of mitochondria of trypanosomes.
PROCESSING OF r-RNA:
Processing of prokaryotic r-RNA:
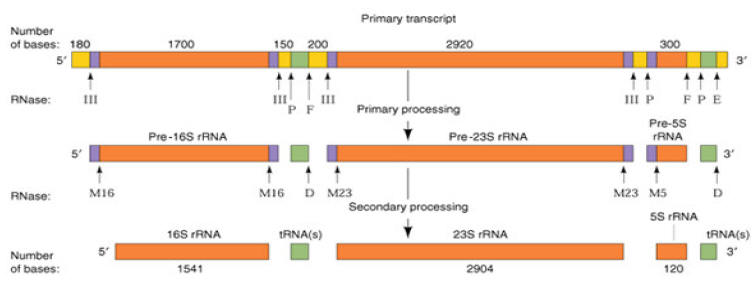
The prokaryotic r-RNAs are of three types namely
a) 16S r-RNA (1541 ribonucleotides)
b) 23S r-RNA (2904 ribonucleotides)
c) 5S r-RNA (120 ribonucleotides)
The primary transcript of these ribosomal RNAs is linked together as a nucleotide chain of more than 5,500 ribonucleotides. The primary r-RNA transcript undergoes processing in the following steps
i) Primary processing
ii) Secondary processing
iii) Methylation
i) Primary processing:
In this process, the primary rRNA undergoes cleavages in which the rRNAs (16S, 23S and 5S) and tRNAs are cleaved by trimming of the flanking nucleotide sequences. The trimming involves endonucleolytic cleavage which is catalyzed by RNase III, RNase P, RNase E and RNase F.
ii) Secondary processing:
The endonucleolytic activity of RNse III, P, E and F does not completely trim the flanking regions of the r-RNAs. The 5 and 3 ends of 16S r-RNA, 23S r-RNA and 5S r-RNA gets further trimmed by the RNase M16, M23 and M5 respectively and RNase D involves in the trimming of flanking regions of t-RNA. After secondary processing, the rRNAs get associated with proteins to form ribosomes.
iii) Methylation:
During ribosomal assembly, the 16S r-RNA and 23S r-RNA are methylated at a total of 24 specific nucleoside residues. The methylation reaction which employ S-adenosine methionine, a methyl donor, yield N6, N6 Dimethyl adenine and 2-o-methyl ribose residues which are thought to protect adjacent phosphodiester bond from degradation by intracellular RNases. This is because RNases hydrolysis involves utilization of the free 2-OH groups of ribose. However, the function of base methylation is unknown.
Processing of Eukaryotic r-RNA:
The eukaryotic r-RNAs are of four types namely
a) 18S r-RNA (1900 nucleotides)
b) 5.8S r-RNA (160 nucleotides)
c) 28S r-RNA (4700 nucleotides)
d) 5S r-RNA (120 nucleotides)
The Primary r-RNA transcript is of approximately 13,000 nucleotide residues and has a sedimentation coefficient of 45S. Starting from 5-end, the structural arrangement of various r-RNAs in the pre-r-RNA is as follows:
5 --- 18S r-RNA ---- 5.8S r-RNA ----28S r-RNA ----3
As in prokaryotes, these r-RNAs separated by spacer sequences. Processing of these r-RNAs involved in the following steps
i) Methylation
ii) Primary processing
iii) Secondary processing
iv) Splicing
i) Methylation:
In the first stage of its processing, 45S r-RNA is specifically methylated at approximately 110 sites that occur mostly in its r-RNA sequences. About 80% of these modifications yield O2 methyl ribose residues and the remainder form methylated bases such as N6, N6 Dimethyladenine and 2-methylguanine. After, methylation, pre r-RNA undergoes other stages of processing.
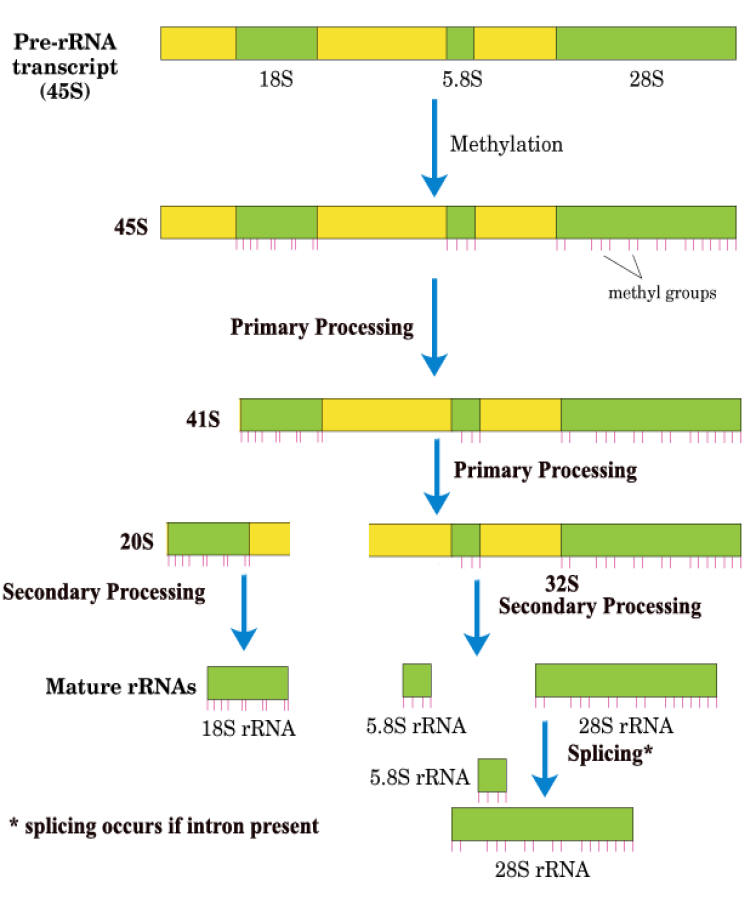
ii) Primary processing:
After methylation, 45S r-RNA undergoes cleavage of 5-end spacer to yield 41S r-RNA. The next step involves cleaving 41S r-RNA into two pieces, 32S and 20S that contains the 28S and 18S sequences respectively. The 32S precursor also retains the 5.8S RNA sequence. This ends primary processing stage.
iii) Secondary processing:
In this stage, 32S precursor is split to yield the mature 28S and 5.8S RNAs, which base pair with each other and the 20S precursor is trimmed to mature 18S size.
iv) Splicing:
Only a few eukaryotic r-RNA genes contain introns. So, they alone undergo splicing.
Example:
26S part of the protozoan Tetrahymena thermophilia r-RNA precursor does contain introns and it can be spliced by 26S r-RNA itself without any help from proteins. The Tetrahymena 26S r-RNA is the equivalent of the mammalian 28S r-RNA.
Mechanism of splicing:
Group I introns splicing:
Splicing of 26S r-RNA examined and explained by Thomas Cech. The introns in 26S r-RNA of Tetrahymena are known as group I introns which also occur in the nuclei, mitochondria and chloroplast of diverse eukaryotes (although not vertebrates). In the first step of splicing, a guanine nucleotide attacks the adenine nucleotide residue at the 5-end of the introns, releasing exon-1 from the rest of the molecule and leaving introns -1 and exon -2 complexes.
In the second step, exon-1 attacks exon-2, performing the splice reaction that releases linear introns and joins the two exons together.
Group II introns splicing:
Introns of yeast mitochondrial pre-r-RNA are known as group-II introns which also occur in the mitochondria of fungi and plants and comprise the majority of the introns in chloroplasts. Group II introns also self splice but they do not need assistance from guanosine to start the reaction. Instead, the initiating entity is an adenosine nucleotide residue within the introns of the RNA itself.
In the first step, 2-OH of adenosine residue attacks the 5-end nucleotide residue to form lariat structure with introns-1 and exon-2 complex and release exon-1.
In the second step, exon-1 attacks exon-2, performing splice reaction tat release lariat introns and joins the two exons together.
The details of this r-RNA processing scheme are not universal. Even the mouse does things a little differently and the frog precursor is only 40S which is quite a bit smaller than 45S. Still, the basic mechanism of r-RNA processing, including the order of mature sequences in the precursor is preserved throughout the eukaryotic kingdom.
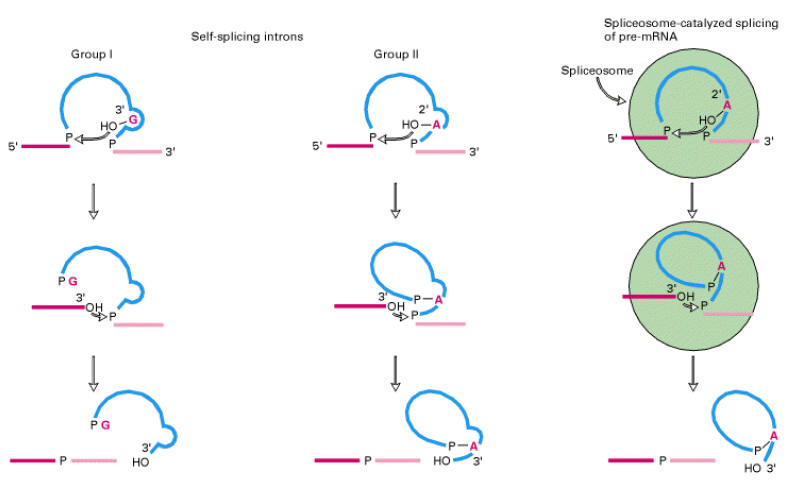
Ribozymes:
RNAs with enzymatic activities are referred as ribozymes. Example: Hammerhead ribozymes of plant virus and Tetrahymena thermophilia r-RNA. Since splicing carried out by RNA itself, the process is known as self splicing.
Processing of t-RNA:
Both prokaryotic and eukaryotic pre t-RNA undergo post transcriptional modification. The steps for post transcriptional modification of pre-t-RNA are as follows:
i) First, the flanking regions of the 3-OH and 5 phosphate ends are cleaved by the endonuclease action of RNase D and RNase P respectively.
ii) Then, the introns in the anticodon loop space, spliced out by splicing reaction.
iii) Tri nucleotide CCA is added to the 3-end to give 32-OH ACC terminus. This reaction is catalyzed by t-RNA specific nucleotidyl transferase. It is unique reaction because this enzyme catalyzes transfer of 3 nucleoside phosphodiester bond formation in one step.
iv) Finally, the t-RNA undergoes base modifications to give mature t-RNA.
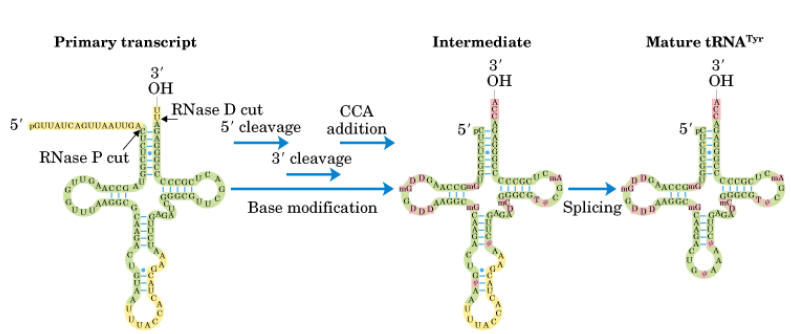
Splicing mechanism:
Splicing mechanism in pre-t-RNA differ from mechanisms utilized by self splicing introns and spliceosomes.
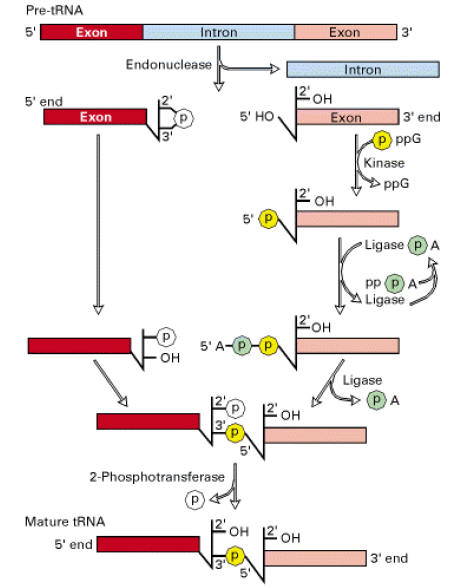
Splicing reactions requires four enzymes namely t-RNA specific endonuclease, cyclic phosphodiesterase, t-RNA specific ligase and 2-phosphotransferase.
By the action of endonuclease, introns removed. Following, excision of introns, a 2-3 cyclic phosphomonoester bond forms on the cleaved end of the 5-exon. The multi step reaction joining the two exon requires two nucleoside triphosphates: a GTP, which contributes the phosphate group for the 3ΰ5 linkage in the finished t-RNA molecule and an ATP, which forms an activated ligaseAMP intermediate. The 2 phosphate on the 5-exon is removed in the final step.
INHIBITORS OF RNA METABOLISM:
A large variety of inhibitors of RNA synthesis have been identified. The inhibitors fall into three groups. They being
i) Inhibitors act by binding to DNA
ii) Inhibitors act by binding to RNA polymerase
iii) Inhibitors act by binding to RNA chain
i) Inhibitors act by binding to DNA:
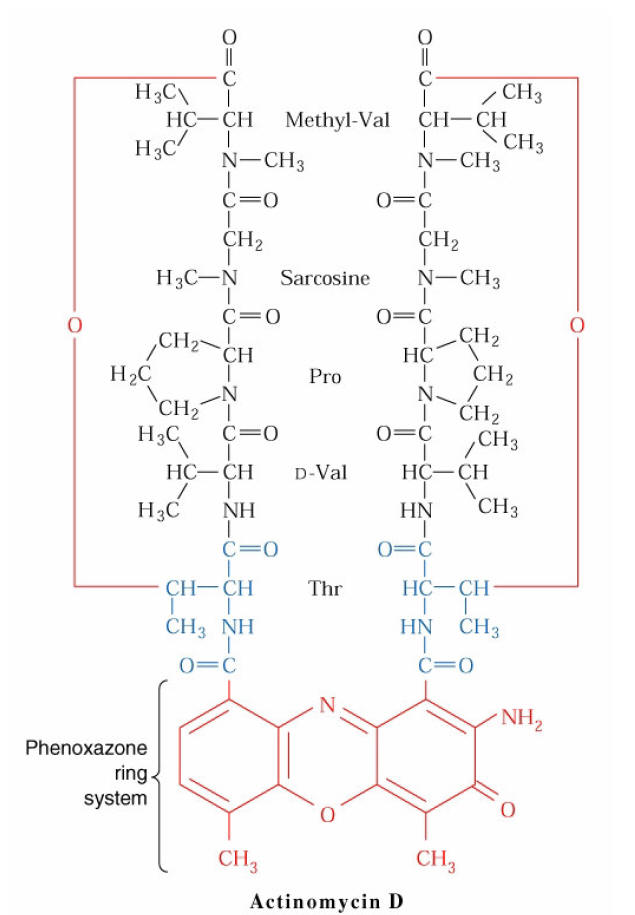
The best known example of inhibitors that bind to DNA is Actinomycin D, an antibiotic produced by streptomyces antiboticus. The inhibition of RNA synthesis is caused by the insertion (interaction) of its phenoxazone ring between two G-C pairs, with the side chains projecting into the minor groove of the double helix, hydrogen bonded to guanosine residues. RNA Polymerase binding to DNA that contains Actinomycin D is only slightly impaired, but RNA chain elongation in both eukaryotes and prokaryotes is blocked.
Ethidium bromide also intercalates into DNA and at low concentrations preferentially binds to negatively supercoiled DNA. It has been used to selectively inhibit transcription in mitochondria which contains supercoiled DNA.
ii) Inhibitors act by binding to RNA Polymerase:
Rifampicin is a synthetic derivative of a naturally occurring antibiotic, Rifampicin that inhibits bacterial DNA dependent RNA polymerase but not T7 RNA polymerase or eukaryotic RNA polymerase. It binds tightly to the beta subunit. Although it does not prevent promoter binding or formation of the first phosphodiester bond, it effectively prevents synthesis of longer RNA chains. It does not inhibit elongation when added after initiation has occurred.
Another antibiotic, streptolydigin, also binds to the beta subunit, it inhibits all bond formation.
The most useful inhibitors of eukaryotic transcription have been a-amanitin, a major toxic substance in the poisonous mushroom Amanita phalloides. The toxin preferentially binds to and inhibits RNA Pol-II. At high concentrations it also can inhibit RNA Pol-III but not RNA Pol-III but not RNA Pol-I or bacterial, mitochondrial or chloroplast RNA Polymerases.
iii) Inhibitors act by binding to growing RNA chain:
Cordycepin in its 5-triphosphorylated form is a substrate analog that is incorporated into growing RNA chains by most RNA polymerases. It causes chain termination after incorporation, since it does not contain the 3-hydroxyl group necessary for the formation of the next phosphodiester bond.
Inhibition like Nalidixic acid, Novobiocin and Dichloro ribo benzene (DRB) etc., also inhibit transcription.
VIRUSES:
Depending upon the genomic nature, it may be classified into three types namely DNA viruses, RNA viruses and Prions.
DNA viruses:
DNA viruses have DNA as a genetic material. Three strategies are used by different DNA viruses to accomplish transcription of viral DNA.
The first type, utilizes the host RNA Polymerases, in some cases modifying it or synthesizing new promoter specific factors to direct to read the viral promoters. Ex: Bacteriophage f 174, T4.
The second type of virus utilizes the host RNA Polymerase to transcribe early viral genes including a gene for a new RNA Polymerase that transcribes exclusively the remaining late viral genes. E.Coli bacteriophage T1 and T3 are examples of this type.
A third type of virus, exemplified by bacteriophage N4, carries a virus specific RNA polymerase in its virion. This polymerase enters the cell together with the viral DNA and transcribes some early viral genes. Some of these genes code for specificity factors that direct the host RNA Polymerase to transcribe late genes. Vaccinia virus is another example of a virus that contains a virion encapsulated RNA Polymerase.
RNA Viruses:
There are mainly two types of RNA viruses namely RNA replicase containing and Reverse transcriptase containing viruses.
RNA viruses containing RNA replicase:
This is further grouped into two types. They are positive strand virus and negative strand virus.
Positive strand viruses:
Viruses like QB, MS2, R17 and f2 contain t-RNA strand which itself act as m-RNAs. Bacteriophage QB codes for a polypeptide that combines with three host proteins to form an RNA dependent RNA Polymerase (RNA replicase). The three host proteins are ribosomal proteins S1 and two elongation factors for protein synthesis: EF-TU and EF-TS.
The QB replicase functions exclusively with the QB RNA plus stand template. It first makes a complementary RNA transcript (minus strand) and ultimately uses the minus strand as a template to synthesize multiple copies of viral RNA plus strands.
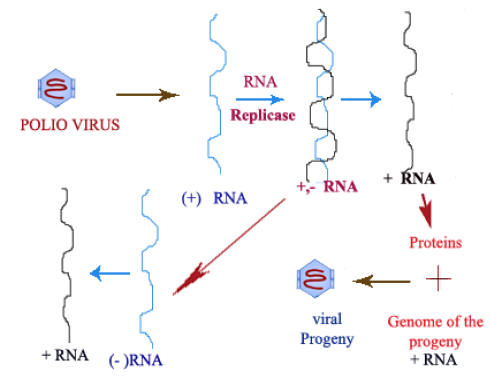
Like the DNA dependent RNA Polymerases, the RNA replicases utilize ribonucleotides and transcribes in the 5ΰ3 direction. The phage RNA must first act as an m-RNA to direct the synthesis of the aforementioned component of the replicase. Since uninfected cells do not have an RNA dependent RNA Polymerase or RNA replicase.
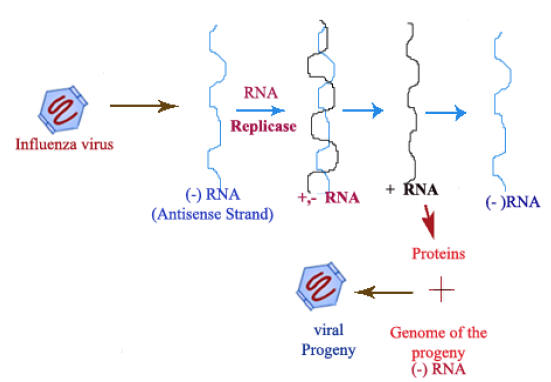
Negative Strand Viruses:
Viuses like Influenza virus contain minus strand RNA as a genetic material. It is an antisense stand. RNA replicase acts on minus RNA stand to form + stands. + strands are used to synthesize viral proteins for progeny formation. They also used to form minus strands which acts as genome of the progeny.
RNA Viruses containing Reverse Transcriptase:
Viruses like human Immuno deficiency virus (HIV), the Rous Sarcoma Virus, feline leukemia virus and mouse mammary tumor viruses are examples of RNA viruses containing reverse transcriptase. They are retroviruses which are a group of animal viruses named for their backward (retro) way of replicating their nucleic acids. The unique feature of the retroviruses is their ability to produce DNA from RNA.
The RNA molecules have associated with it a copy of the enzyme reverse transcriptase. This enzyme is so named because it uses the genomic RNA as template to synthesize DNA. This enzyme first discovered independently by David Baltimore and Howard M. Temin in 1970. This enzyme is RNA directed DNA Polymerase which has three enzyme activities namely
i) RNA directed DNA Polymerase
ii) Ribonuclease H activity
iii) DNA directed DNA Polymerase
i) RNA directed DNA Polymerase:
By using RNA directed DNA Polymerase activity, reverse transcriptase produces a single stranded DNA molecule using RNA as the template.
ii) Ribonuclease H activity
H- refers to hybrid. By using ribonuclease it activity, so DNA freed from RNA-DNA hybrid.
iii) DNA directed DNA Polymerase:
By means of DNA directed DNA Polymerase activity, dsDNA synthesized from ssDNA.
After dsDNA formed, it get incorporated into host cell genome and it gets expressed along with host genome and viral progeny produced.
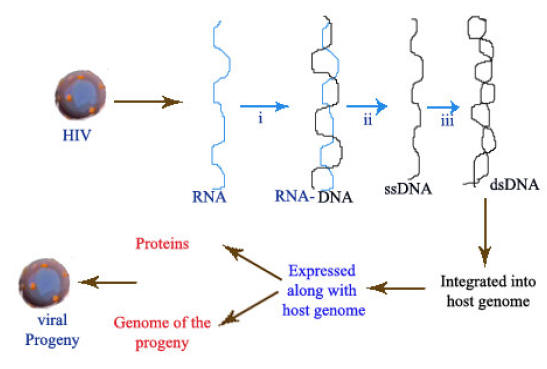
Reverse Transcriptase as Biomedical Tool:
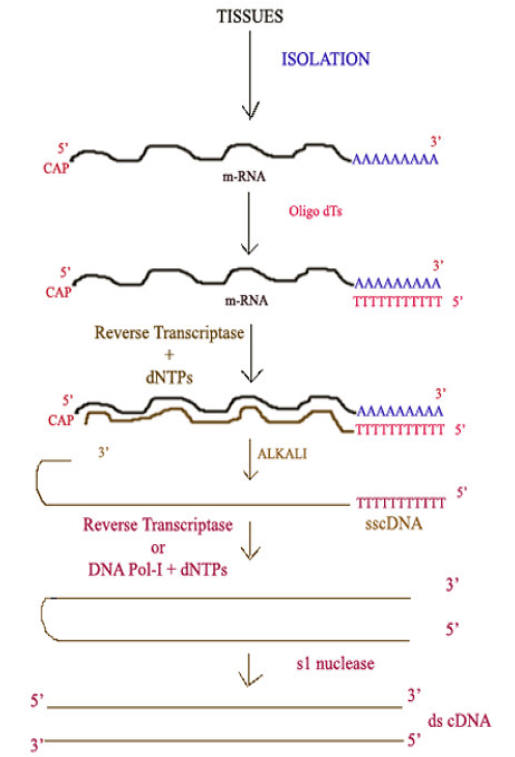
Reverse Transcriptase becomes a powerful biomedical tool in genetic engineering. It is used in cloning DNA. Reverse Transcriptase makes possible laboratory synthesis of DNA, complementary in base sequence to an RNA template. A synthetic DNA prepared in this manner is called complementary DNA [cDNA], which is then cloned with bacterial plasmid and introduced into a bacterial culture which then gives rise to cDNA library.
Synthesis of cDNA:
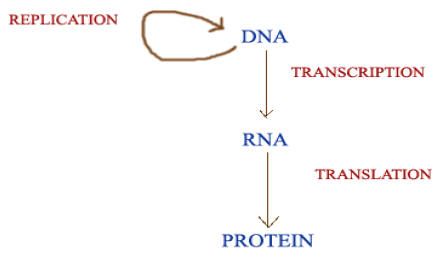
CENTRAL DOGMA OF MOLECULAR BIOLOGY:
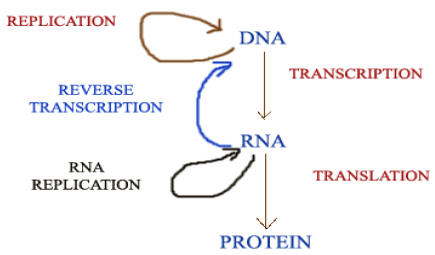
Central dogma of Molecular Biology states that the genetic information can flow from DNA to DNA, DNA to RNA and RNA to protein only. It is represented as below:
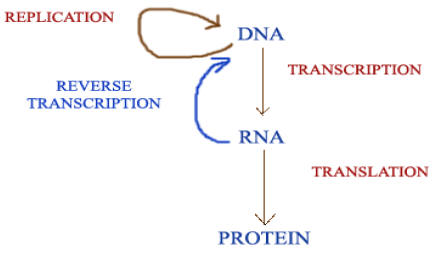
MODIFIED CENTRAL DOGMA:
The discovery of reverse transcriptase has modified the central dogma of molecular biology which held that genetic information should pass only from DNA to RNA. This enzyme, synthesis DNA forms RNA, thus showing that the information can flow from RNA to DNA.
Like reverse transcriptase, RNA replicase also has modified the central dogma molecular biology. Normally, information transferred from DNA to RNA by Transcription and RNA to DNA by reverse transcriptase but RNA replicase transfer the information from RNA to RNA. Thus it modifies the dogma of molecular biology. Modification by RNA replicase is as follows: