REPLICATION IN PROKARYOTES
Replication is an enzymatic process in which synthesis of a daughter or progeny duplex DNA molecule, identical to the parental duplex DNA occurs. Rate of replication in E.Coli (prokaryotic cell) is 1500 nucleotides per second. To complete replication of whole E.Coli genome it takes 40 minutes. Rate of replication in eukaryotes is about 10 - 100 nucleotides per second. To complete replication of simple eukaryotic genome 6 - 8 hours required. In prokaryotic circular DNA only one replication fork is present but in eukaryotic DNA several replication forks are present. Space between two-replication forks in eukaryotes is about 20kbps apart.
THE REPLICATION OF CIRCULAR DNA IN E.COLI (Prokaryotic duplex DNA replication) (q REPLICATION):
The synthesis or replication of DNA molecule can be divided into three stages
I) Initiation (Formation of Replisome)
II) Elongation (Initiation of synthesis and elongation)
III) Termination
I) Initiation
The replication begins at a specific initiation point called OriC point or replicon. (Replicon: It is a unit of the genome in which DNA is replicated; it contains an origin for initiation of replication) It is the point of DNA open up and form open complex leading to the formation of prepriming complex to initiate replication process.

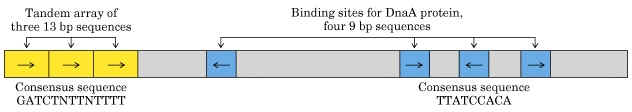
The OriC site is situated at 74" minute near the ilv gene. The OriC site consists of 245 basepairs, of which three of 13 basepair sequence are highly conserved in many bacteria and forms the consensus sequences (GATCTNTTNTTTT). Close to OriC site, there are four of 9 basepair sequences each (TTATCCACA).
The sequence of reactions in the initiation process is as follows:
a) Dna A protein recognizes and binds up to four 9bp repeats in OriC to form a complex of negatively supercoiled OriC DNA wrapped around a central core of Dna A protein monomers. This process requires the presence of the histone like HU or 1 HC proteins to facility DNA bending.
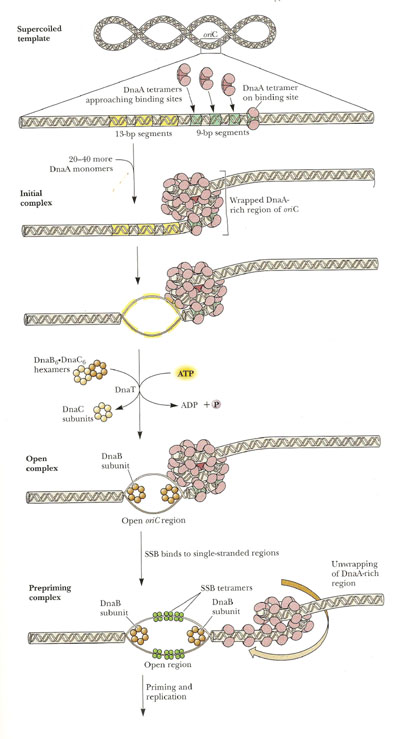
b) Dna A protein subunits then successively melt three tandemly repeated 13bp segments in the presence of ATP at >=22*C (open complex).
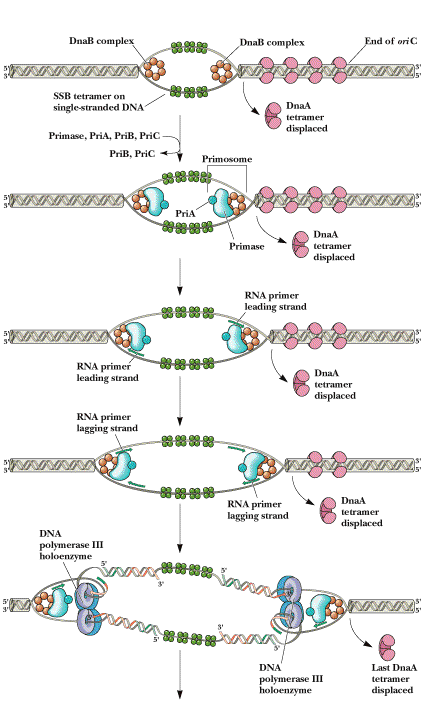
c) The Dna A protein then guides a Dna B - Dna C complex into the melted region to form a so called prepriming complex. The Dna C is subsequently released. Dna B further unwinds open complex to form prepriming complex.
d) DNA gyrase, single stranded binding protein (SSB), Rep protein and Helicase - II are bound to prepriming complex and now complex is called as priming complex.
e) In the presence of gyrase and SSB, helicases further unwinds the DNA in both directions so as to permit entry of primase and RNA polymerase. Then RNA polymerase forms primer for leading strand synthesis while primase in the form of primosome synthesis primer for lagging strand synthesis.
f) To the above complex, DNA polymerase - III will bind and forms replisome.
REPLISOME: It is the multiprotein structure that assembles at the bacterial replicating fork to undertake synthesis of DNA. It contains DNA polymerase and other enzymes.
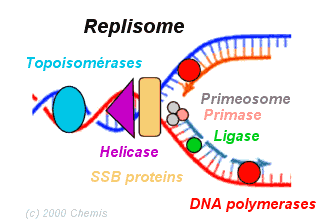
II) ELONGATION:
Now the stage is set for the initiation of synthesis and the elongation to proceed. But this occurs in two mechanistically different pathways in the 5'-->3' template strand and 3'-->5' template strand.
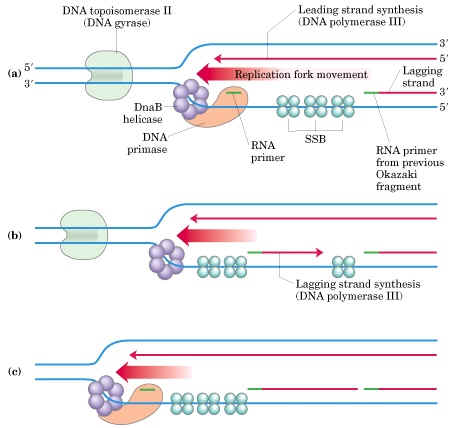
a) Initiation of synthesis and Elongation on the 5'-->3' template (synthesis of leading strand) (If replication fork moves in 3'-->5' direction)
The DNA daughter strand that is synthesized continuously on 5'-->3' template is called leading strand. DNA pol-III synthesizes DNA by adding 5'-P of deoxynucleotide to 3'-OH group of the already presenting fragment. Thus chain grows in 5'-->3' direction. The reaction catalyzed by DNA pol-III is very fast. The enzyme is much more active than DNA pol - I and can add 9000 nucleotides per minute at 37*C. The RNA primer that was initially added by RNA polymerase is degraded by RNase.
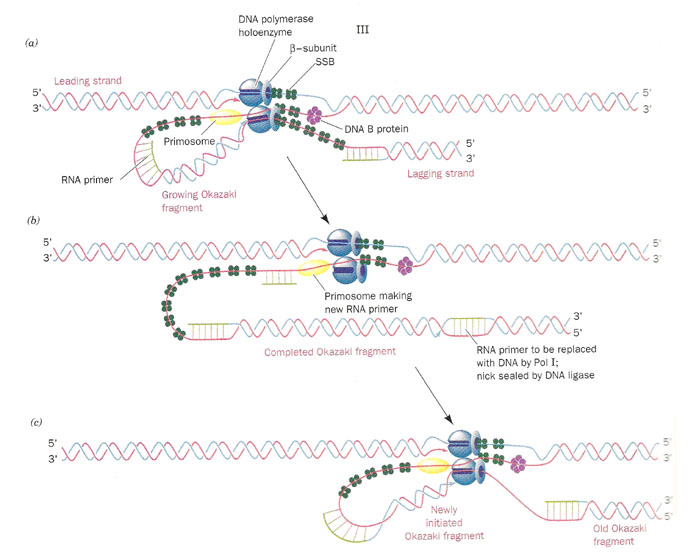
b) Initiation of synthesis and Elongation on 3'-->5' template when fork moves in 3'-->5' direction (Synthesis of lagging strand)
The daughter DNA strand which is synthesized in discontinuous complex fashion on the 3'-->5' template is called lagging strand. It occurs in the following steps:
i) Synthesis of Okazaki fragment:
To the RNA primer synthesized by primosome, 1000-2000 nucleotides are added by DNA pol-III to synthesis Okazaki fragments.
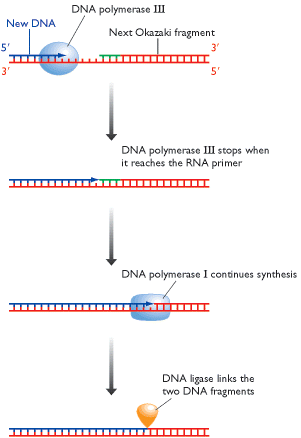
ii) Excision of RNA primer:
When the Okazaki fragment synthesis was completed up to RNA primer, then RNA primer was removed by DNA pol - I using its 5'-->3' exonuclease activity.
iii) Filling the gap (Nick translation)
The gap created by the removal of primer, is filled up by DNA pol - I using the 3'-OH of nearby Okazaki fragment by its polymerizing activity.
iv) Joining of Okazaki fragment: (Nick sealing)
Finally, the nick existing between the fragments are sealed by DNA ligase which catalyze the formation of phosphodiester bond between a 3'-OH at the end of one strand and a 5' - phosphate at the other end of another fragment. The enzyme requires NAD for during this reaction.
III) TERMINATION:
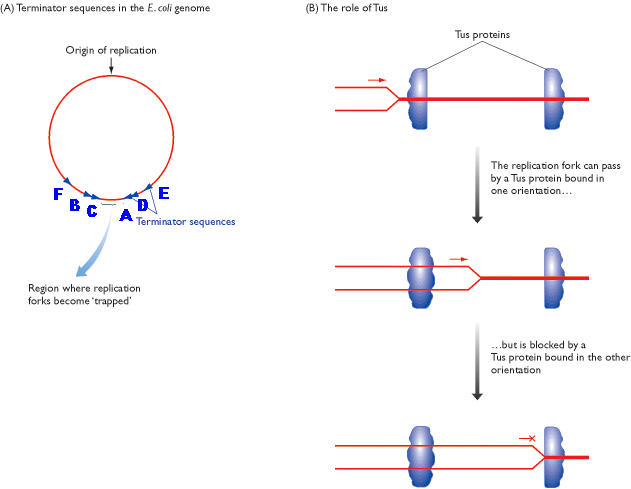
Termination occurs when the two replicating forks meet each other on the opposite side of circular E.Coli DNA. Termination sites like A, B, C, D, E and F are found to present in DNA. Of these sites, Ter A terminates the counter clockwise moving fork while ter C terminates the clockwise moving forks. The other sites are backup sites. Termination at these sites are possible because, at these sites tus protein (Termination utilizing substance) will bound to Dna B protein and inhibits its helicase activity. And Dna B protein released and termination result.
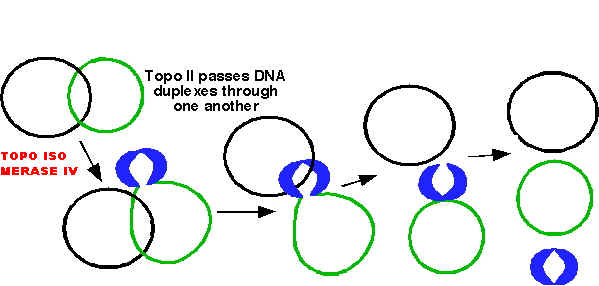
After the complete synthesis, two duplex DNA are found to be catenated (knotted). This catenation removed by the action of topoisomerase. Finally, from single parental duplex DNA, two progeny duplex DNA synthesized.
REGULATION OF PROKARYOTIC REPLICATION:
Especially initiation of replication is regulated. Dna A protein when available in high concentration then ratio of DNA to cell mass is quiet high but at low Dna A concentration, the ratio found to be low. This shows that Dna A protein regulates the initiation of replication.
The sequence most commonly methylated in E.Coli is GATC including in three of 13mer sequence. Thus, the observation that E.Coli defective in the GATC methylation enzyme are very inefficiently replicated, suggests that the DNA replication trigger also responds to the level of OriC methylation.
OTHER MODELS FOR CIRCULAR DNA REPLICATION:
There are two such models are available namely Rolling Circle and D-loop models.
i) Rolling circle model OR s-replication:
In the rolling circle model of replication, a nick is made in one of the strands of the circular DNA, resulting in replication of circle and a tail. This form of replication occurs in the F plasmid or E.Coli Hfr chromosome during conjugation. The F+ or Hfr cell retains the circular daughter while passing the linear tail into the F- cell, where replication of tail takes place. This method is also used in several phages (Viruses), which fill their heads with linear DNA replicated form a circular parent molecule.
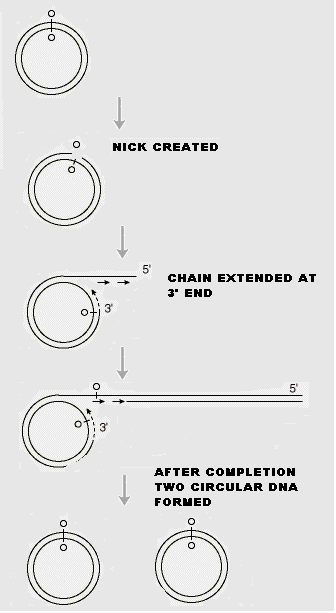
In some cases, because there is no termination point, synthesis often continues beyond a single circle unit, producing concatamers i.e., a series of linked chains, of several circle lengths, which are then processed by recombination to yield normal length circles.
ii) Displacement loop or D-Loop model:
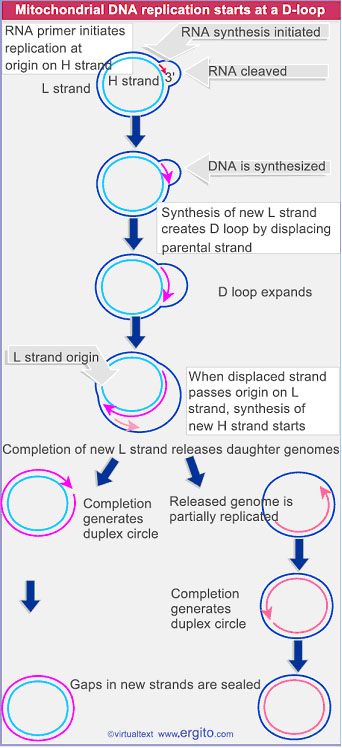
Chloroplasts and mitochondria in eukaryotic cell have their own circular DNA molecules that appear to replicate by a slightly different mechanism than those described. The origin of replication is at a different point on each of the two parental template strands. Replication begins on one strand, displacing the other while forming a displacement loop or D-loop structure. Replication continues until the process passes the origin of replication on the other strand. The newly synthesized strand, known as leading strand. Replication is then initiated on the second strand in the opposite direction which is the lagging strand. When leading strand completely replicated, only 1/3 of lagging strand is replicated. Then finally the result is two circles.
f or Fi X174 BACTERIOPHAGE REPLICATION:
f X174 Bacteriophage contains a small single-stranded circular DNA. It occurs in two stages namely formation of Replicative Form and formation of '+' strand.
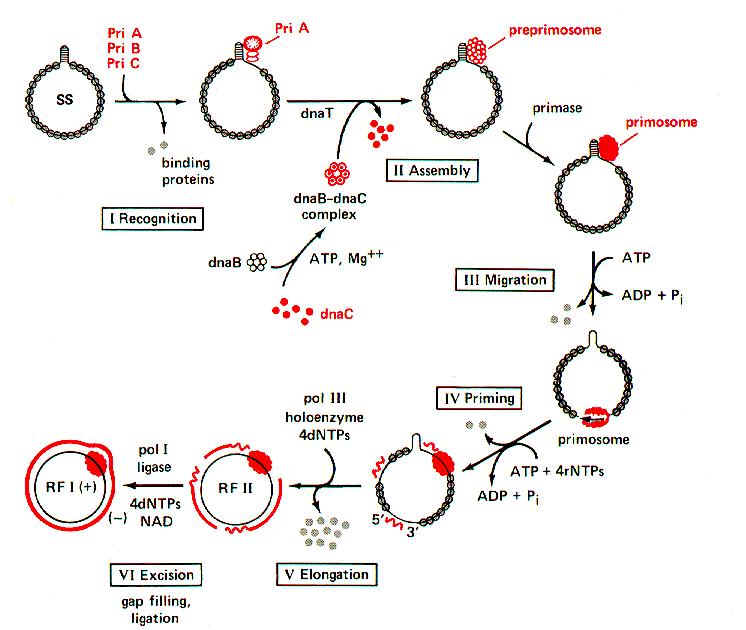
i) Synthesis of '-' strand or Replicative Form I (RFI):
It occurs in six steps. They are as follows:
a) To the (+) strand, PriA, PriB and PriC (formerly known as n', n, and n'' respectively) bound to the pas (Primosome assembly site) site. It contains 70 nucleotides hairpin. It is present between genes F and G.
b) Dna B and Dna C complex then bound to the DNA with the help of Dna T protein (formerly known as 'i') in an ATP - requiring process. Dna C then released and forming the preprimosome complex. When Primase bound to the preprimosome complex, it is referred as primosome.
c) Primosome complex moves in 5’-->3' direction on DNA with the help of ATP hydrolysis. Pri A and Dna B aid this function.
d) During the movement on the '+' DNA strand Primase synthesis RNA Primer. This function also requires Dna B protein to maintain the DNA template conformation.
e) Then DNA pol III extends the primers to form Okazaki fragments.
f) DNA pol I then removes the primers and replaces them with DNA. The nicks are then joined by DNA ligase and supercoiled by DNA gyrase. Thus RF I form of f X174 formed. The primosome remains complexed with '+' DNA even in RF I form.
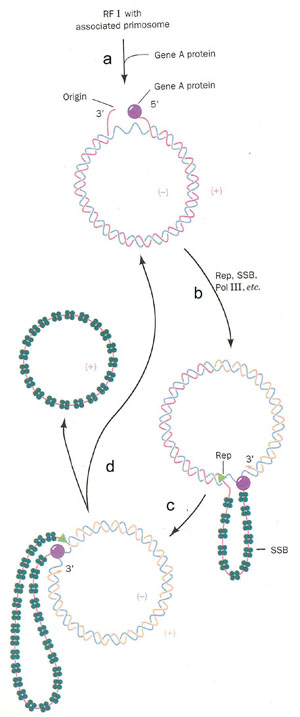
ii) Synthesis of '+' Strand or Looped Rolling Circle model or s Model:
It occurs in four steps. They are as follows:
a) Gene A protein bound to the 30bp region near the beginning of gene A in RF I with the primosome aiding function. Gene A cleaves the phosphodiester bond and forms a covalent bond between a Tyr residue and the DNA's 5’-phosphoryl group. This reaction is used to conserve the energy present in the phosphodiester bond.
b) Rep protein binds to (-) strand and commences unwinding the duplex DNA form the 5' end. The binding is aided by the primosome complex. The displaced (+) strand is coated with SSB protein which prevents the recoiling of (+) strand with (-) strand. Rep protein is essential for fX 174 replication unlike E.Coli replication. DNA pol - III then extends the (+) strand from 3' - end.
c) The extension process generates a looped rolling circle structure in which the 5'end of the old (+) strand remains linked too the gene A protein at the replication fork. Primosome found to be attached with the old (+) strand which may used for later generation of fX 174.
d) After full circle of (-) strand replicated, Gene A protein make a cut in the (+) strand at the replication fork and joins the old (+) strands and new (+) strands. Because of this, the SSB bound old (+) strand released. At the end of this step, RF I form regenerated and one (+) strand DNA synthesized. RF I is used for the production of more (+) strand and viral proteins where as (+) strand utilized for the formation of new phages.
EUKARYOTIC DNA REPLICATION (Replication of Linear DNA):
Eukaryotic replication occurs during s-phase of cell cycle. Replication usually occurs only one time in a cell. Replication in eukaryotes occur in five stages namely,
1. Pre-initiation:
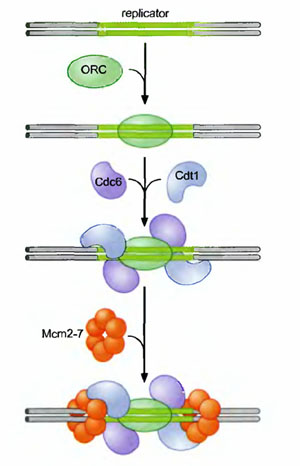
Actually during pre-initiation stage, replicator selection occurs. Replicator selection is the process of identifying the sequences that will direct the initiation of replication and occur in G1 phase. and occurs in Gl (prior to S phase). This process leads to the assembly of a multiprotein complex at each replicator in the genome. Origin activation only occurs after cells enter S phase and triggers the Replicator - associated protein complex to initiate DNA unwinding and DNA polymerase recruitment. Replicator selection is mediated by the formation of pre-replicative complexes (pre-RCs). The first step in the formation of the pre-RC is the recognition of the replicator by the eukaryotic initiator, ORC (Origin recognition Complex). Once ORC is bound, it recruits two helicase loading proteins (Cdc6 and Cdtl). Together, ORC and the loading proteins recruit a protein that is thought to be the eukaryotic replication fork helicase (the Mem 2-7 complex). Formation of the pre-RC does not lead to the immediate unwinding of origin DNA or the recruitment of DNA polymerases. Instead the pre-RCs that are formed during Gl are only activated to initiate replication after cells pass from the Gl to the S phase of the cell cycle.
2. Initiation:
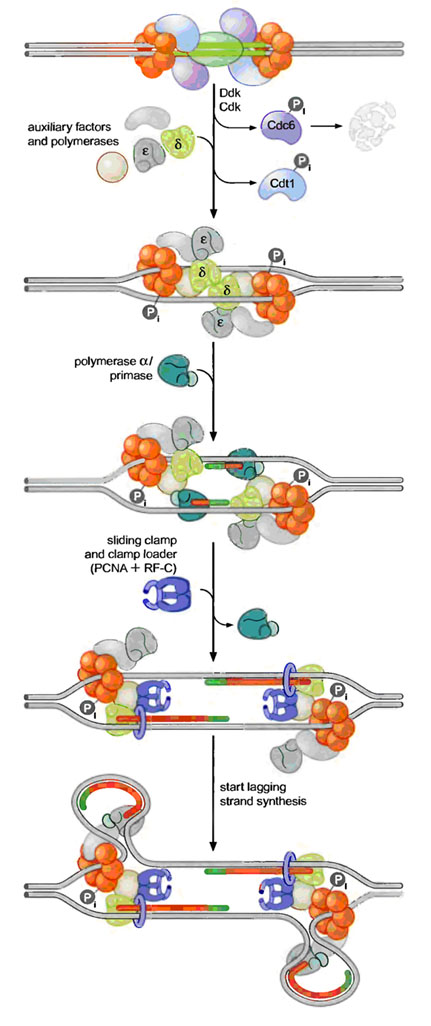
Pre-RCs are activated to initiate replication by two protein kinases namely Cdk (Cyclin Dependant Kinase) and Ddk (Ddt4 Dependant Kinase). Kinases are proteins that covalently attach phosphate groups to target proteins. Each of these kinases is inactive in Gl and is activated only when cells enter S phase. Once activated, these kinases target the pre-RC and other replication proteins. Phosphorylation of these pro-proteins results in the assembly of additional replication proteins at the origin and the initiation of replication.
These new proteins include the three eukaryotic DNA polymerases and a number of other proteins required for their recruitment. Interestingly, the polymerases assemble at the origin in a particular order. DNA Pol d and e associate first, followed by DNA Pol a/primase. This order ensures that all three DNA polymerases are present at the origin prior to the synthesis of the first RNA primer (by DNA Pol a/primase). Once present at the origin, DNA Pol a/primase synthesizes an RNA primer and briefly extends it. Thus initiation of replication started.
3. Elongation:
The resulting primer-template junction is recognized by the eukaryotic sliding clamp loader (RF-C), which assembles a sliding clamp (PCNA) at these sites. Either DNA Pol d or e recognizes this primer and begins leading strand synthesis. After a period of DNA unwinding, DNA Pol a/primase synthesizes additional primers, which allow the initiation of lagging strand DNA synthesis by either DNA Pol d or e. In the diagram, Pol d was used for leading strand and Pol e was used for lagging strand synthesis. DNA Pol e possess activity to remove primer and fills the gap with DNA like DNA Pol I in prokaryotes. SSB like activity was played by replication protein A (RP A) which is denoted as accessory factors during replication.
4. Termination:
When the replication forks meet each other, then termination occurs. It will result in the formation of two duplex DNA. Eventhough replication terminated, 5’ end of telomeric part of the newly synthesized DNA found to have shorter DNA strand than the template parent strand. This shortage corrected by the action of telomerase enzyme and then only the actual replication completed.
5. Terlomerase Function:

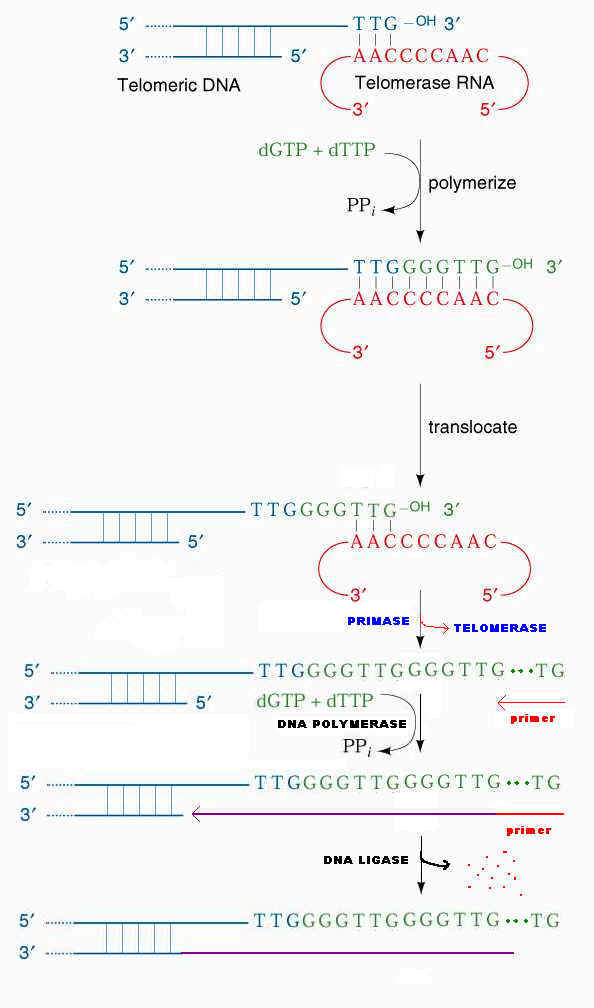
In Linear eukaryotic chromosome, once the first primer on each strand is remove, then it appears that there is no way to fill in the gaps, since DNA cannot be extended in the 3'-->5' direction and there is no 3' end upstream available as there would be in a circle DNA. If this were actually the situation, the DNA strand would get shorter every time they replicated and genes would be lost forever.
Elizabeth Blackburn and her colleagues have provided the answer to fill up the gaps with the help of enzyme telomerase. So, that the genes at the ends, are conserved. Telomerase is a ribonucleoprotein (RNP) i.e. it has RNA with repetitive sequence. Repetitive sequence varies depending upon the species example Tetrahymena thermophilia RNA has AACCCC sequence and in Oxytrica it has AAAACCCC. Telomerase otherwise known as modified Reverse Transcriptase. In human, the RNA template contains AAUCCC repeats. This enzyme was also known as telomere terminal transferase.
The 3'-end of the lagging strand template basepairs with a unique region of the telomerase associated RNA. Hybridization facilitated by the match between the sequence at the 3'-end of telomere and the sequence at the 3'-end of the RNA. The telomerase catalytic site then adds deoxy ribonucleotides using RNA molecule as a template, this reverse transcription proceeds to position 35 of the RNA template. Telomerase then translocates to the new 3'-end by pairing with RNA template and it continues reverse transcription. When the G-rich strand sufficiently long, Primase can make an RNA primer, complementary to the 3'-end of the telomere's G-rich strand. DNA polymerase uses the newly made primer to prime synthesis of DNA to fill in the remaining gap on the progeny DNA. The primer is removed and the nick between fragments sealed by DNA ligase.
REGULATION OF EUKARYOTIC REPLICATION:
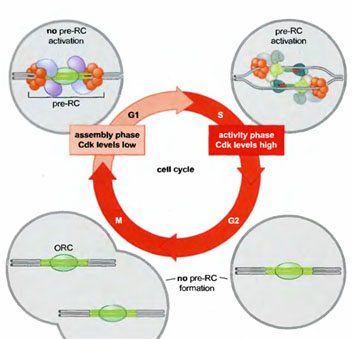
The tight connection between pre-RC function, Cdk levels, and the cell cycle ensures that the eukaryotic genome is replicated only once per cell cycle (Figure 8-32). Active Cdk is absent during Gl, whereas elevated levels of Cdk are present during the remainder of the cell cycle (S, G2, and M phases). Thus, during each cell cycle there is only one opportunity for pre-RCs to form during Gl and only one opportunity for those pre-RCs to be activated during S, G2, and M phase, although in practice all pre-RCs are activated or disrupted by replication forks in S phase. Pre-RCs are disassembled after they are activated or after the DNA to which they are bound is replicated. These exposed replicators are then available for new pre-RC formation and rapidly bind to ORC. Despite the presence of the initiator at these sites, the elevated levels of Cdk activity in S, G2, and M phase cells prevents the association of the other members of the pre-RC complex with ORC. It is only when cells segregate their chromosomes and complete cell division that Cdk activity is eliminated and new pre-RC complexes can form.
Inhibition of replication was achieved by antisense RNA. Once antisense RNA produced for particular gene, it bound with single strand DNA in open complex and inhibits the movement of replisome and thus replication. Antisense RNA synthesized in opposite direction to the normal RNA synthesis.
INHIBITORS OF REPLICATION:
Dauromycin and Adriamycin:
They are synthetic chemotherapeutic agents and are inhibitors of both DNA replication and transcription in prokaryotes. These are presumably act by interfering with the passage of both DNA and RNA polymerase. They have planar aromatic ring system which gets intercalated between GC pairs of the double helical structure. Thus, they prevent its replication and transcription.
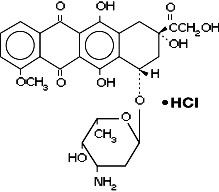
Adriamycin is otherwise known as doxorubicin. Doxorubicin is a cytotoxic anthracycline antibiotic isolated from cultures of Streptomyces peucetius var. caesius. Doxorubicin consists of a naphthacenequinone nucleus linked through a glycosidic bond at ring atom 7 to an amino sugar, daunosamine. Doxorubicin binds to nucleic acids, presumably by specific intercalation of the planar anthracycline nucleus with the DNA double helix. The anthracycline ring is lipophilic, but the saturated end of the ring system contains abundant hydroxyl groups adjacent to the amino sugar, producing a hydrophilic center. The molecule is amphoteric, containing acidic functions in the ring phenolic groups and a basic function in the sugar amino group. It binds to cell membranes as well as plasma proteins.
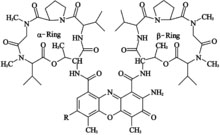
Actinomycin - D
It is an antibiotic produced by streptomyces and inhibits replication and transcription. It acts by intercalating its phenoxazone ring between two successive GC pairs in duplex DNA. Actinomycin D has two identical pentapeptides which have unusual composition of D-Valine and Sarcosine which stabilizes this intercalating interaction.
It was the first antibiotic shown to have anti-cancer activity, but is not normally used as such, as it is highly toxic, causing damage to genetic material. Actinomycin-D is marketed under the trade name Dactinomycin. Actinomycin-D is one of the older chemotherapy drugs which has been used in therapy for many years. It is a clear, yellow liquid which is administered intravenously and most commonly used in treatment of a variety of cancers.
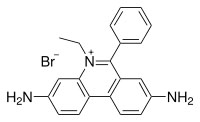
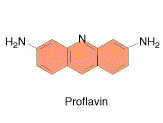
Ethidium bromide and proflavin
They inhibit both replication and transcription by intercalation. As with most fluorescent compounds, it is aromatic. The main portion of the molecule is a tricyclic structure with aniline (aminobenzene) groups on either side of a pyridine (a six-atom, nitrogen-containing, aromatic ring). The dibenzopyridine structure is known as a phenanthridine. The reason for ethidium bromide's intense fluorescence after binding with DNA is probably not due to rigid stabilization of the phenyl moiety, because the phenyl ring has been shown to project outside the intercalated bases. In fact, the phenyl group is found to be almost perpendicular to the plane of the ring system, as it rotates about its single bond to find a position where it will abut the ring system minimally. Instead, the hydrophobic environment found between the base pairs is believed to be responsible. By moving into this hydrophobic environment and away from the solvent, the ethidium cation is forced to shed any water molecules that were associated with it. As water is a highly efficient fluorescent quencher, the removal of these water molecules allows the ethidium to fluoresce. This property is used to identify the presence of DNA in gel und UV light.
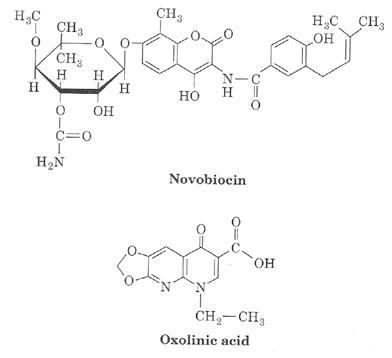
Novobiocin and oxolinic Acid
Prokaryotic DNA gyrases are specifically inhibited by two classes of antibiotics. One of these classes includes the streptomyces-derived novobiocin and the other contains the clinically useful synthetic antibacterial agent oxolinic acid.
Both classes of antibiotics profoundly inhibit bacterial DNA replication and transcription, thereby demonstrating the importance of properly supercoiled DNA in these processes. Studies using antibiotic-resistant E.Coli mutants have demonstrated that oxolinic acid associates with DNA gyrase’s A subunit and novobiocin binds to its B subunit. When DNA gyrase is incubated with DNA and oxolinic acid, and subsequently denatured with guanidinium chloride, it’s A subunits remain covalently linked to the 5’-ends of both cut strands through phosphotyrosine linkages. Apparently oxolinic acid interferes with gyrase action by blocking the strand breaking-rejoining process. Novobiocin, on the other hand, prevents ATP from binding to the enzyme.
Aphidicolin
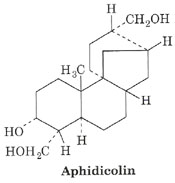
Aphidicolin inhibits DNA pol a of eukaryotes, so it inhibits eukaryotic replication. Aphidicolin also inhibits DNA pol d and DNA pol e. Thus Aphidicolin inhibits both leading and lagging strand synthesis in eukaryotes.
Rifamycin
Rifammycin inhibits RNA polymerase, so RNA primer for leading strand synthesis is not available. Thus replication inhibited. The rifamycins are a group of antibiotics which are synthesized either naturally by the bacterium Amycolatopsis mediterranei, or artificially. The rifamycin group includes the "classic" rifamycin drugs as well as the rifamycin derivatives Rifampicin, Rifabutin and Rifapentine. The biological activity of rifamycins relies on the inhibition of DNA-dependent RNA synthesis. This is due to the high affinity of rifamycins to prokaryotic RNA polymerase. Crystal structure data of the antibiotic bound to RNA polymerase indicates that rifamycin blocks synthesis by causing strong steric clashes with the growing oligonucleotide. If rifamycin binds the polymerase after the chain elongation process has started, no effect is observed on the biosynthesis, which is consistent with a model that suggests rifamycin physically blocks the chain elongation. In addition, rifamycins showed potency towards tumors. This is due to their inhibition of the enzyme reverse transcriptase, which is essential for tumor persistence. However, rifamycins potency proved to be mild and this never leads to their introduction to clinical trials.
FIDELITY OF REPLICATION:
It refers to the precised state or accuracy of replication. The rate of error occurring in E.Coli is one mispairing per 108 to 1010 base pairs replicated. This corresponds to one error per 1000 bacteria per generation. There are four main reasons for this high level of accuracy in replication. They are as follows:
a) Cells maintain balanced level of dNTPs, thus each base added correctly to the binding site of the polymerase enzyme. But if one present at low levels then it is more likely to be replaced by the dNTPs present at higher levels.
b) The catalytic mechanism of action of polymerase itself responsible for high fidelity. This is because polymerization reaction occurs in two stages namely binding and catalysis. In the binding step, nucleotide binds to binding site in the enzyme and it forms Watson-crick base pair with the base in the template and then phosphodiester bond forms between the bases after it forms complementary base pairing. Binding process actually checks the binding of the correct base by checking the base pairing.
c) The 3'-->5' exonuclease activity of DNA POL I and DNA POL III also detect and eliminate the errors made by their polymerase function during replication.
d) Normal repair system present in all cells also plays a vital role in the maintenance of DNA.
In addition to the above four major reasons for the fidelity, there are two minor reasons also present. They are Primer requirement and Lagging strand synthesis. Due to cooperative nature of base pairing, first few nucleotides might be added incorrectly. But due to the presence of primers, this error does not occur because primer replaced by the DNA later correctly. If DNA polymerase synthesizes DNA in 3'-->5' direction also, then when wrong base pair is added during proof reading function, then polymerase introduce the 5'-OH or 5' - Phosphate terminal region, and then the extension of the end is not possible without reactivation. Thus by forming lagging strand the end extension is possible and proof reading function also occurs normally so it prevents unusual base pairing during replication.
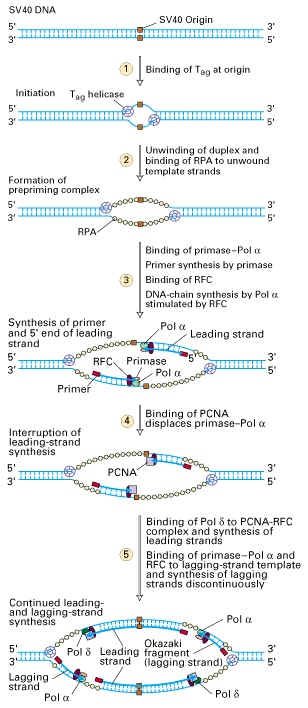
SV 40 VIRAL REPLICATION:
Replication initiated by the binding of T antigen (Virus encoded protein). This multifunctional protein then unwinds DNA through its helicase activity. This reaction requires ATP. RF A (Replication Factor A), host cell protein then binds to single strands and stabilize it similar to SSB. To the open complex, pol a - Primase complex binds and Primase forms RNA primer. This primer elongated by pol a using deoxy nucleotides whose activity is stimulated by RF C. PCNA binds to the primer template terminus and displaces pol a - Primase complex. Pol d then binds to PCNA at the 3' - ends of other growing strand and continues the leading strand synthesis. PCNA was responsible for the processivity of pol d. As unwinding of the duplex DNA progresses further away from the origin, the Primase -pol a complex associates with the unwound template strands downstream from the leading strand primers. Synthesis of lagging strand then is carried out by primase - pol a and RF C. Finally, topoisomerase probably play an important role in relieving tortional stress induced by growing fork movement and in separating the two daughter duplex DNA.