MODES OF REPLICATION
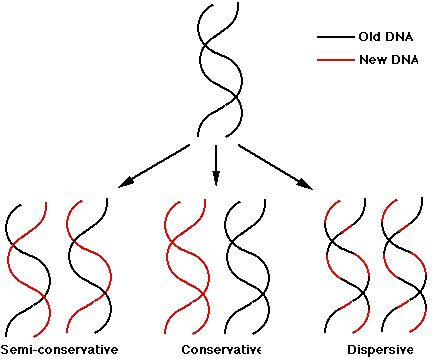
There are three modes are proposed for replication by Crick namely Conservative mode, Semiconservative mode and Dispersive mode. Of the three, Semiconservative mode of replication in prokaryotes proved by Meselson-Stahl experiment.
WATSON AND CRICK MODEL OR HYPOTHESIS ON REPLICATION:
The Watson and Crick model for DNA replication assumes that as new strands of DNA are made, they follows the usual base pairing rules of A with T and G with C. This is essential because the DNA replicating machinery must be capable of discerning a good pair from a bad one, and the Watson - Crick base pairs give the best fit. Three hypothesis available for DNA replication. They are
i) Conservative replication
ii) Semiconservative replication
iii) Dispersive replication

Conservative replication: It yields two daughter duplexes, one of which has two old strands and one of which has two new strands.
Semiconservative replication: It gives two daughter duplex DNAs, each of which contains one old strand and one new strand.
Dispersive replication: It yields two daughter duplexes, each of which contains strands that are mixture of old and new DNA.
EVIDENCE FOR SEMI CONSERVATIVE REPLICATION:
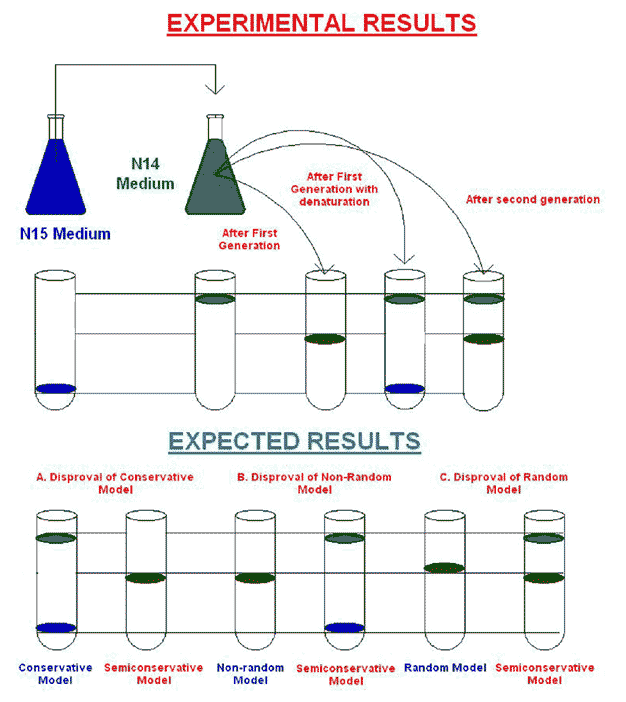
In 1958, Matthew Meselson and Franklin Stahl worked out clever procedure to distinguish semi conservative DNA replication from conservative or dispersive replication, using a nonradioactive heavy isotope of nitrogen. Ordinary nitrogen, the most abundant isotope, has an atomic weight of 14, so it is called 14N. A relatively rare isotope 15N has an atomic weight of 15. Meselson and Stahl found that if bacteria grow in a medium enriched in 15N, they incorporate the heavy isotope into their DNA, which becomes denser than normal. This labeled DNA CLEARLY separates from ordinary DNA in gradient of Cesium Chloride (CsCl) spun in an ultracentrifuge. CsCl is used because it is a very dense salt and therefore makes dense enough solution that DNA will float somewhere in the middle rather than sinking to the bottom.
AIM:
The crux or aim of the experiment was to grow 15N labeled bacteria in 14N-medium for one or more generations and then to look at the density of the DNA products.
RULING OUT CONSERVATIVE MODEL:
Consider the different outcomes we would expect after one round of replication according to the three different mechanisms. If replication is conservative, the two heavy parental strands will stay together and another, newly made DNA duplex will appear. Since this second duplex will be made in the presence of light isotope, both its strands will be light. The heavy / heavy parental duplex and light / light progeny duplex will separate readily in the CsCl gradient. On the other hand, if replication is semi conservative or dispersive, the two heavy parental stands will separate and each will be supplied with a new light partner and heavy and light strand dispersed randomly between parental and progeny strands respectively. These heavy / light hybrid duplexes will have a density halfway between the heavy / heavy parental DNA and light / light ordinary DNA. Meselson & Stahl results confirmed the latter prediction, so the conservative mechanism ruled out.
RULING OUT RANDOM DISPERSIVE MODEL:
The results of one more round of DNA replication destroyed the random dispersive hypothesis. The random dispersive model predicts that the DNA after the second generation will have a single density, corresponding to molecules that are 25% heavy and 75% light. This should give one band of DNA halfway between light / light band and the heavy / light band. The semi conservative model predicts equal amounts of two different DNAs. Again, the latter prediction matched the experimental results, confirming the semi conservative model.
RULING OUT NON-RANDOM DISPERSIVE MODEL:
Nonrandom dispersive distribution would yield the same gradient centrifugation result as would semi conservative replication after two rounds of replication. To eliminate the possibility of such dispersive replication, Meselson and Stahl took DNA after one round of replication and heated it to separate its stands. Then they subjected the separated strands to gradient centrifugation. The semi conservative model predicts that half the strands should be heavy and half should be light. These should form separate bands upon centrifugation coincident with standards prepared from pure heavy and light DNAs. On the other hand, any dispersive model predicts that both stands should be mixed and therefore should not form bands coincident with pure heavy and light DNA strands.
In Meselson and Stahl's experiment, the two strands were readily separable, and behaved completely light and completely heavy DNA strands, just as the semi conservative model demands. This allows us to rule out the nonrandom dispersive hypothesis.
CONCLUSION:
DNA replicates in a semi conservative manner. When the parental strands separate, each serves as the template for making a new, complementary strand.
AUTORADIOGRAPHIC DEMONSTRATION OF DNA REPLICATION (John Cairns Experiment):
The Semiconservative method of replication was verified photographically in 1963 by J.Cairns who used the technique of autoradiography. This technique makes use of the fact that radioactive atoms expose photographic film. The visible silver grains on the film can then be counted to provide an estimate of the quantity of radioactive material present. Cairns grew E.Coli bacteria in a medium containing radioactive thymine, a component of one of the DNA nucleotides. The radioactivity was in tritium (31H). The DNA was then carefully extracted from the bacteria and placed on photographic emulsion for a period of time. The emulsion was then developed to produce autoradiograph that was examined under the electron microscope. Each grain of silver represents a radioactive decay. Interpretation of this autoradiograph reveals several points.
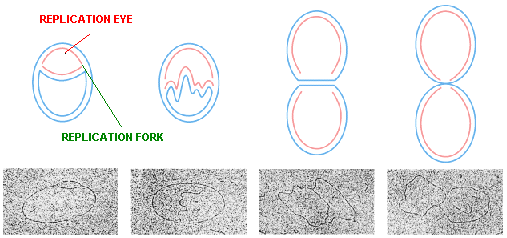
The first, known at the time, is that the E.Coli DNA is a circle. The Second point is that DNA is replicated while maintaining the integrity of the circle i.e., the circle does not appear to be broken in the process of DNA replication; an intermediate theta structure is formed which is due to the formation of replication eye. Third, replication of the DNA seems to be occurring at one or two moving Y-junctions in the circle Replication forks, which further supports the Semiconservative replication.
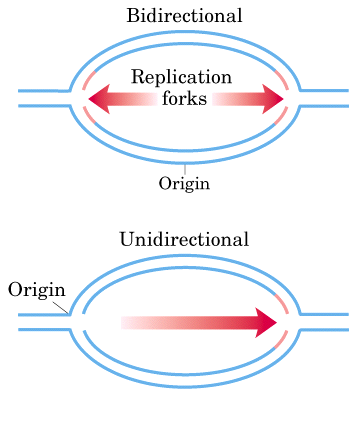
REPLICATION IS BI-DIRECTIONAL:
In some cases, radioactivity had not been applied to the cell until DNA replication had already begun. In these cases, the radioactive label appeared after the theta structure had already begun forming. After some time, the process was stopped and the autoradiographs, Cairns found growth to be bi-directional. This finding has subsequently been verified by both autoradiographic and genetic analysis.
Result:
As the grain track pattern present on both forks with unlabeled DNA in the middle, it was concluded that the direction of DNA synthesis is bi-directional.
The unidirectional replication was ruled out because had it been the case, the grain tracks should have been on only one side of the point of origin.
REIJI OKAZAKI EXPERIMENT (Semidiscontinuous Replication):
Autoradiograph images of replicating DNA of E.Coli suggest that DNA's two antiparallel strands are simultaneously replicated as the replicating fork advances in diametrically opposite directions.
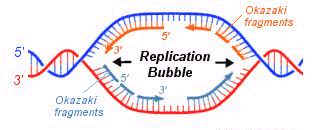
All known DNA polymerases catalyze the chain growth only in 5'--> 3' direction. Therefore, synthesizing daughter DNA on 3’--> 5' template is done by polymerase as the 3' group is free for the chain growth in 5'-->3' direction. But how can polymerase catalyze replication on template of 3'-->5'. In this case only 5' end would be available, polymerase not use it to start the chain growth. Reiji Okazaki’s Pulse-Chase Experiment answered this baffling question.
Okazaki's model of Semidiscontinuous replication makes two predictions that his research team tested experimentally.
1) Since atleast half of the newly synthesized DNA appears first as short pieces, it is necessary to fix the short pieces of DNA by labeling them with radioactive precursors. These short bursts of labeling with radioactive substances are called pulses. The stopping period is called chase.
2) If we eliminate the enzyme (DNA ligase) that is responsible for Stiching together the short pieces of DNA, we ought to too be able to see these pieces even with long pulses of DNA precursor.
For his model system, Okazaki chases replication of bacteriophage T4 DNA. This had the advantage of simplicity, as well as the availability of mutants of T4 DNA ligase.
To test first prediction, Okazaki gave shorter and shorter pulses of tritium-labeled thymidine to E.Coli cells that were replicating T4 DNA (as short as two seconds). Finally, he measured the approximate size of the newly synthesized DNAs by ultracentrifugation. It is found to be between 7S - 11S. Large DNA pieces sediment faster than smaller pieces toward the bottom of the centrifuge tube. The smaller DNA fragments found to have 1000-2000 nucleotides in length. With increasing pulse time, another band of tritium labeled DNA appears much nearer to the bottom of centrifuge tube. This is the result of attaching small, newly formed pieces of labeled DNA too much larger. These fragments are referred as Okazaki fragments.
Okazaki's group performed second experiment with the T4 mutant containing a defective DNA ligase group. In this case, Okazaki fragments found to be present for even one minute of pulses. Therefore Reiji Okazaki's Pulse Chase experiment with the replicating T4m DNA molecules showed that the DNA replication in one of the strand is occurs in a Semidiscontinuous fashion.
REPLICATION EYE:
It is formed during replication in both eukaryotic and prokaryotic DNA. It is the place where replication occurs actively. It is otherwise known as replication bubble. Formation of the replication eye provides the theta like structure to the circular DNA during replication in prokaryotes. Each replication bubble found to have two replication forks, each at the corner of an eye.
REPLICATION FORK:
The corner region of the replication bubble or replication eye is referred as replication fork. Generally it is point from which the replication elongated or continued. Bi-directional nature of replication was identified by studying the appearance of nucleotides at the replication forks in connection with the change in the radioactive material intensity. Each replication bubble found to have two replication forks, each at the corner of an eye.
SEMIDISCONTINUOUS REPLICATION:
During Replication, half of the DNA strand synthesized in fragments where as remaining half synthesized continuously. This is referred as semidiscontinuous replication. This happens only because of the nature of the DNA polymerases i.e., they can synthesize DNA by extending 3’end and they are unable to extend DNA from 5’end. This nature of replication is proved by Okazaki by an experiment.
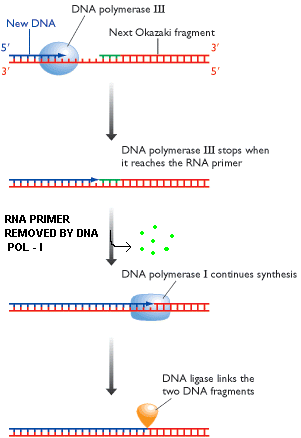
OKAZAKI FRAGMENTS:
Fragments synthesized during lagging strand formation of replication was identified and proved by Rejis Okazaki. Hence, by his name these fragments are called as Okazaki’s fragments. They are short polynucleotides with 1000-2000 base pairs in length. These fragments are synthesized by DNA polymerases. Even though they are formed during replication, they are joined to form larger DNA at the completion of replication by the action of DNA ligase. The invention of Okazaki fragments lead to the proposal of semidiscontinuous replication concept.

RNA PRIMERS:
RNA primers are single strand oligoribonucleotides with 40 to 60 base pairs. For replication RNA primers are required. This is because DNA polymerases require RNA primer for their action to start in invivo condition. In replication RNA primer is synthesized by two different enzymes namely RNA polymerase and Primosome complex. RNA polymerase synthesize RNA primer for synthesize of leading strand where as Primosome synthesize RNA primer for lagging strand synthesis. In addition to this, RNA polymerase synthesizes only one primer whereas Primosome synthesizes many primers.
ENZYMES OF REPLICATION:
DNA DIRECTED DNA POLYMERASES:
This enzyme synthesizes DNA by utilizing DNA template.
PROKARYOTIC DNA DIRECTED DNA POLYMERASES:
In prokaryotes, there are three kinds of DNA polymerases. They are
I. DNA directed DNA polymerase - I
II. DNA directed DNA polymerase - II
III. DNA directed DNA polymerase - III
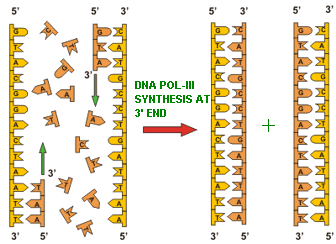
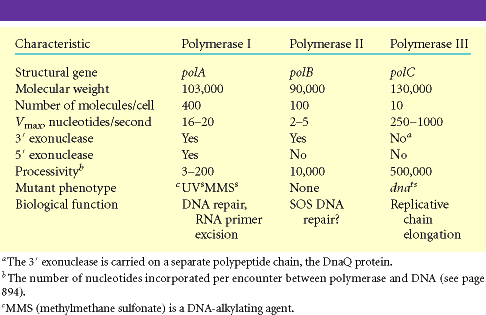
I. DNA directed DNA polymerase - I (Kornberg enzyme):
Its Molecular weight is 109kd and the concentration is around 400 molecules per cell. When treated with proteases like Trypsin, it gets cleaved into two fragments possessing different function. The large fragment called klenow fragment posses both polymerase and 3'-->5' exonuclease activity. The small fragment possesses only 5'-->3' exonuclease activity. Overall, three activities are possessed by DNA Pol-I.
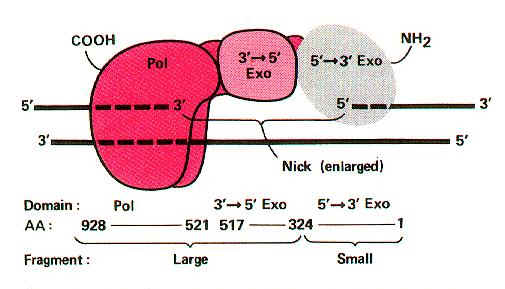
i) DNA polymerase activity
ii) 3'--> 5' exonuclease activity
iii) 5'-->3' exonuclease activity
i) DNA polymerase activity:
It catalyzes synthesis of complementary DNA strand from the appropriate deoxy nucleotides using single or double stranded DNA as template and DNA or RNA as primer. DNA polymerases catalyze polymerization reaction, which involves step-by-step addition of deoxy nucleotides unit to a DNA or RNA primer chain. It is represented by equation
n(dNTPs)--->(dNMP)n
The chain elongation reaction catalyzed by DNA polymerase occurs by means of a nucleophilic attack of the 3'-OH terminus of the primer on the alpha phosphorous atom of the incoming deoxyribonucleotide and releasing pyrophosphate molecule. The subsequent hydrolysis of "ppi" drives the reaction in the forward direction. It is represented in the following figure:
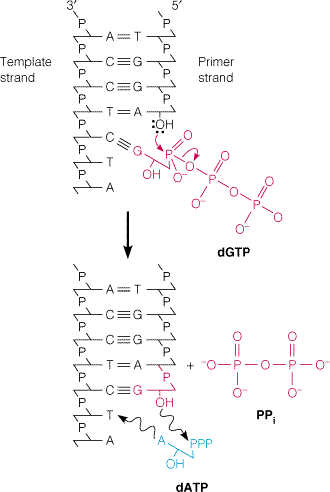
CONDITIONS FOR DNA POLYMERASE ACTION:
a) All four dNTPs (dATP, dGTP, dTTP, dCTP) along with Mg2+ ions.
b) A primer chain with a free 3'-OH group. Invitro conditions require DNA primer strand while invivo condition require RNA primer strand. Generally DNA Pol – I utilizes DNA primer whereas DNA Pol – III utilizes RNA Primer.
c) A DNA template, which can be single stranded or double stranded DNA. Double stranded DNA is an effective template only if its sugar phosphate backbone is broken at one or more sites.
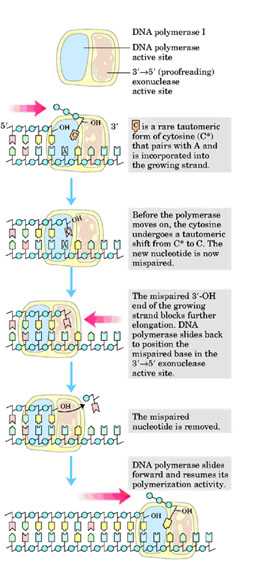
ii) 3'-->5' exonuclease activity:
This is the proofreading or editing activity of DNA polymerase, which gets activated when a wrong base pair is incorporated into the chain. DNA pol-I removes deoxy nucleotide at 3'-end of the progeny DNA. Because of this, the activity is referred as 3'-->5' exonuclease activity. Due to this activity, it repairs DNA during replication.
iii) 5'-->3' exonuclease activity:
With the help of this activity, enzyme removes nucleotides from 5'-end. Two functions can be carried out by this activity. They are removal of RNA primer and DNA repair. Schematic representation for this function is as follows:
a) Removal of Primer:
b) DNA repair:
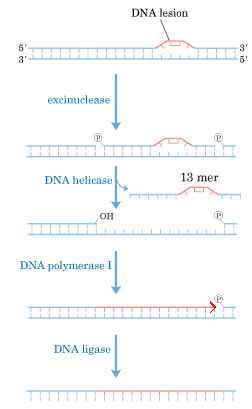
II) DNA directed DNA Polymerase-II:
Structure and function are not completely elucidated. It has polymerase activity along with proof reading function, but it does not have 5'-->3' exonuclease activity.
III) DNA directed DNA Polymerase-III:
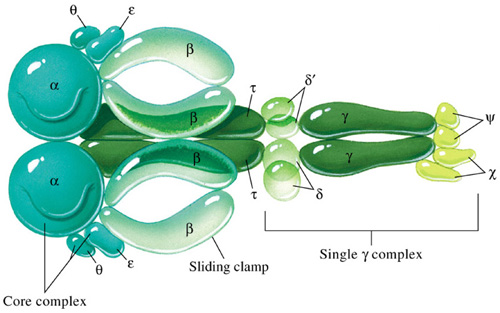
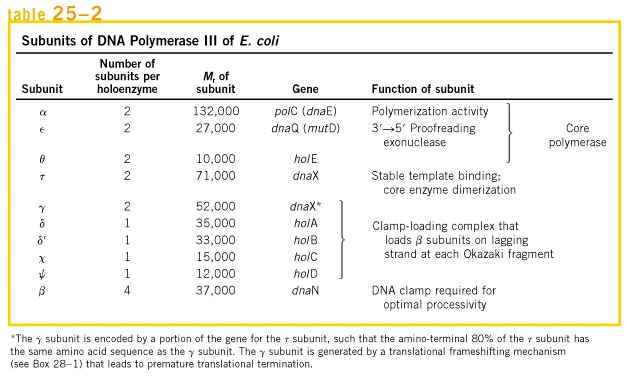
Holoenzyme of DNA pol-III consists of ten subunits. They are aeqbgdd'cjt . Core enzyme of DNA poly-III contain three subunit aeq. Of three subunits alpha subunit posses polymerizing activity and epsilon posses proof reading activity. These are the two function of the enzyme. It does not have 5'-->3' exonuclease activity.
|
S.No |
SUBUNITS |
FUNCTION |
|
1. |
a |
Polymerization |
|
2. |
e |
Repair and Proof reading function |
|
3. |
q |
Assembly of DNA pol-III |
|
4. |
b |
Processivity - It holds the template |
|
5. |
gdd' |
Lagging strand synthesis - They aid the formation of loop. |
|
6. |
cj |
Normal function of DNA pol-III |
|
7. |
t |
Dimerization |
gdd'c and j subunits called as a gamma complex, which mediates transfer of the beta subunit to the duplex DNA primer strand. DNA pol-III possesses high processivity than other DNA polymerases. This is because of beta subunit. It form a donut shaped dimers around duplex DNA and then associate with and hold the catalytic core aeq polymerase at the primer template terminus.
Once tightly associated with DNA, the beta subunit dimers functions like a "clamp" which can move freely along the DNA, like a ring on a string carrying the associated core polymerase with it. In this way, the active sites remain near the growing fork and the processivity of the core polymerase maximized.
Processivity: Number of nucleotides added by the enzyme before it gets released from the template strand after its attachment.
EUKARYOTIC DNA DIRECTED DNA POLYMERASE:
Following are the six important eukaryotic DNA polymerases. They are as follows:
i) DNA polymerase a
ii) DNA polymerase b
iii) DNA polymerase g
iv) DNA polymerase d
v) DNA polymerase e
vi) DNA polymerase s
i) DNA polymerase a
It is composed of four subunits of which one has the primase activity. The largest subunit of it has polymerase activity. It is supposed to exist in several forms like a1,a2, and a3. DNA polymerase a in association with DNA polymerase d , is involved in the replication of nuclear chromosomal DNA. The replication is also aided by another protein factors called Accessory Proteins and AP4A. These protein factors appear to have regulatory function. DNA pol a is supposed to carryout lagging strand synthesis.
ii) DNA polymerase b:
It is involved in the repair of DNA.
iii) DNA polymerase g:
This polymerase is involved in the replication of mitochondria DNA.
iv) DNA polymerase d:
This polymerase has polymerase function and 3'-->5’ exonuclease activity. It is involved in leading strand synthesis.
v) DNA polymerase e:
It is supposed to replace DNA pol delta in some situations such as DNA repair.
vi) DNA polymerase s:
This polymerase seems to be expressed only in the bone marrow cells. It is the only eukaryotic polymerase that has a deoxy ribonuclease activity.
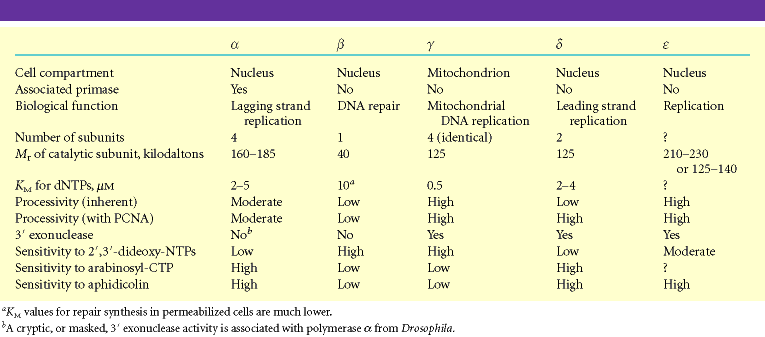
DNA LIGASE:
The DNA ligases are responsible for connecting DNA segments during replication, repair and recombination. They are class of enzymes that catalyze the formation of alpha-phosphodiester bond between two DNA chains. This enzyme requires the free OH group at the 3' end of other DNA strand and phosphate group at 5' end of the other. The formation of a phosphodiester bond between these groups is an endergonic reaction. Hence energy source required for ligation. In E.Coli and other bacteria NAD+ supplies the energy whereas in animals i.e. eukaryotes ATP play the role.
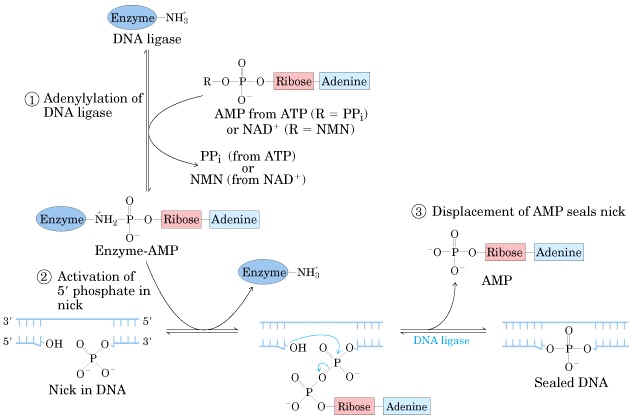
Mechanism:
a) The adenosyl group of the ATP or NAD+ is transferred to the enzyme ligase to form covalent enzyme-AMP complex in which the AMP is linked to the epsilon amino group of lysine residue of the enzyme through posphamide bond.
b) The enzyme then transfers the adenyl group to the 5' phosphoryl terminus of the nick to form a pyrophosphate grouping. In effect, AMP is attached to the 5'-phosphoryl terminus.
c) The final step is a nucleophilic attack of the 3'-OH group on the 5' activated phosphorus atom. A phosphodiester bond is formed and AMP is released. This reaction is driven by the hydrolysis of the pyrophosphate group. Thus two high-energy phosphate bonds are spent in forming a phosphodiester bridge in the DNA backbone.
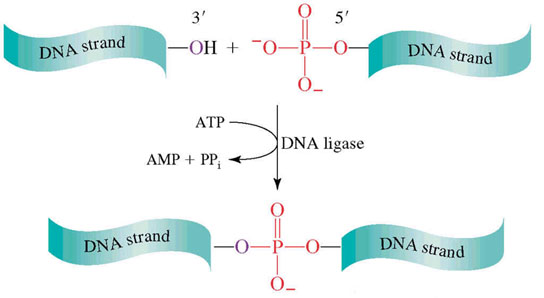
Role of DNA ligases in Replication:
This enzyme seals the nick and joints the Okazaki fragments in the lagging strand. It is also involved in the repair of damaged DNA. DNA ligases are extensively used as a joining tool in genetic engineering.
DNA HELICASES:
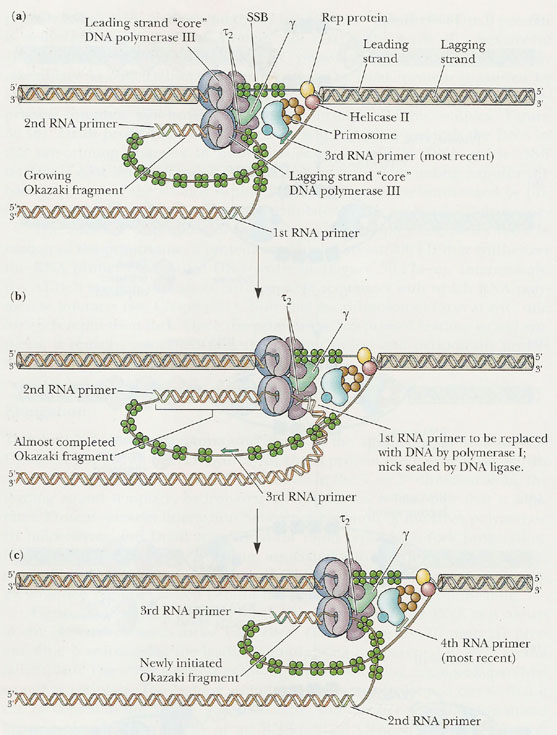
They are proteins, which are involved in the unwinding of DNA molecule. There are four kinds of helicases namely Dna B, Rep Proteins, Helicases - II and Dna A.
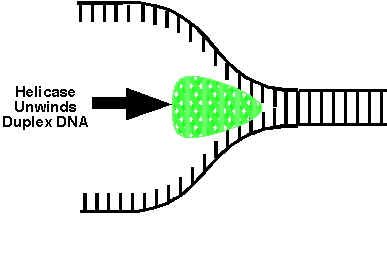
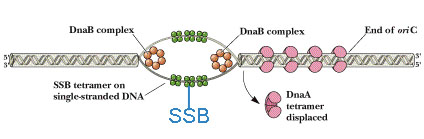
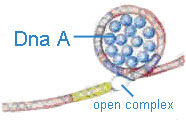
Dna-A protein (Mw 48,000) - It binds to 4 of 9mer sequence and unwinds a 3 of 13mer sequence at Ori C site and forms an open complex during initiation of replication. It is the first protein which binds to DNA to initiate DNA replication.
Dna-B proteins (Mw- 3,00,000)- It is a primosome constituent and consists of six subunits. It unwinds DNA during replication. It is responsible for the extension of open complex during replication.
Rep Proteins (Mw 65,000) - It is a helicase consisting of one subunit. It binds to the 5'3'template and moves in 3'5' direction. It actively participates in leading strand synthesis in replication.
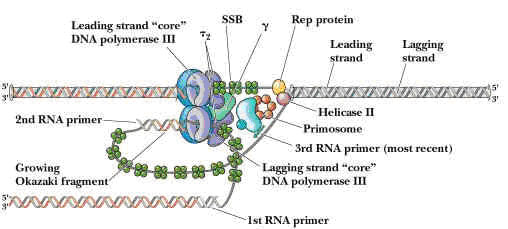
DNA Helicase - II (Mw 75,000) - It is helicase consisting of only one subunit. It binds with 3'5' template stand and moves along in 5'3' direction. It is involved in lagging strand synthesis in replication.
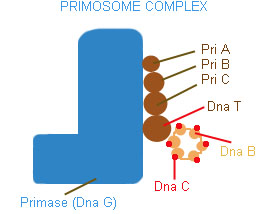
PRIMOSOME:
It is a complex containing Pri A, Pri B, Pri C, Dna T, Dna B, Dna C and Primase (Dna G) proteins. Primososme complex without primase referred as preprimosome complex. Primosome complex plays a vital role in lagging strand synthesis by synthesizing the primer at frequent intervals. Of all the proteins actual primer synthesizing ability relies with primase molecule. But each protein of the complex have their own role i.e. each of them necessary for the function of primosome complex in replication for example. Pri A , Pri B, Pri C necessary for the binding of primosome complex to DNA. Dna B responsible for helicase activity. Dna C aids the binding of Dna B. Dna T helps the binding of DnaB and Dna C complex to Pri A, Pri B and Pri C complex.
SUPERCOILING OF DNA:
When compare to the size of the cells (both in eukaryotes and prokaryotes) the dimensions i.e. the contour length of DNA, is colossally large. The DNA is packed into the cells not by mere folding and stuffing it in a random way but rather in a highly organized fashion as it has to provide an access in a very systematic manner to a vast array of enzymes and other factors that control its replication, transcription and gene regulation. Therefore DNA is accommodated into other small space by an organized bending or twisting of the molecule into a structure called supercoiled structure.
DNA is coiled around an axis in the forms of a double helix and the bending or twisting of that access upon itself is refers to as DNA supercoiling. Therefore, supercoiling represents the twisting or bending of the DNA double helix upon itself. When there is no bending or twisting of DNA axis upon itself, the DNA is said to be in a relaxed state. Supercoiling may occur in two different structural forms, they are intertwined form or toroidal form. In intertwined form, DNA strand twisted upon itself without any support. In toroidal form, it twisted upon a component ex: proteins. Supercoiling of DNA is of two types namely Positive supercoiling and Negative supercoiling.
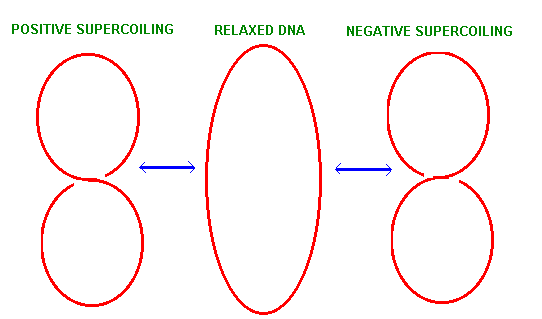
POSITIVIE SUPERCOILING:
If a circular DNA molecule in a relaxed conformation is broken across both strands and one or more additional right handed helical turns are inserted before rejoining the ends. The molecule would twist on itself to form positive super coil.
NEGATIVE SUPERCOILING:
If a circular DNA molecule in a relaxed conformation given a double strand break and unwound by giving left handed helical turn and the ends rejoin, the molecule will twist itself to form negative supercoil.
These two forms of DNA, which will only differ in their topological structures, are called topoisomers.
Terms:
Twist: Number of turns by one-polynucleotide chains around another on double helical axis is referred as twist (t).
Writh: Number of turns by double helical DNA upon itself or around central axis referred as writh (w).
Linking Number: In circular DNA, Linking number (LK) specifies total number of turns that are present in a closed circular DNA. Otherwise, it is a sum of twist and writh.
L= t+ w
Linking Number is a constant property and it can be changed only by phosphodiester bond breaking. If the linking number is reduced by the process of unwinding in the relaxed circular DNA, it induces negative supercoiling. If X is the number of turns in the relaxed circular DNA (Lk0=x) and if one turn is removed by unwinding (LK=X-1), it induces negative supercoiling.
Therefore, the change in linking number DLK= LK - LK0
=X - 1 - X= -1
DLK = -1
On the other hand if one turn is added to the relaxed circular DNA, if result in positive supercoiling, therefore the change in linking number DLK is +1.
TOPOISOMERASES:
The normal biological function of DNA occurs only if it is in proper topological state, i.e. to stay in having proper supercoiling or super helical tension. Topoismerases are a group of enzymes which controls supercoiling of DNA thereby maintaining it in the proper topological state or superhelical tension. There are two classes of topoisomerases. They are type – I topoisomerase and type – II topoisomerase.
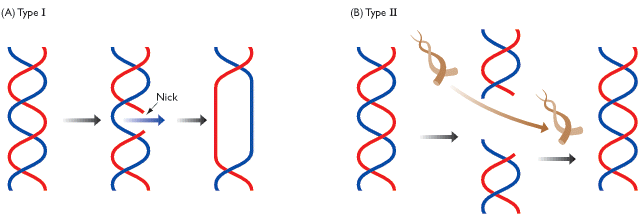
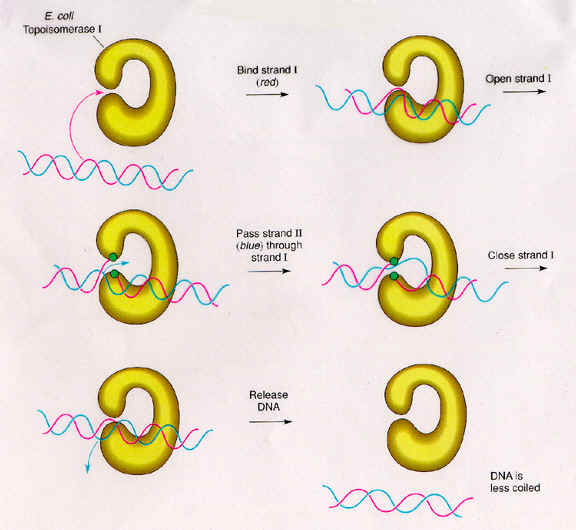
TYPE - I TOPOISOMERASE:
Type-I topoisomerase (Nicking-Closing enzymes) are monomeric 100kd proteins that are widespread in both prokaryotes and eukaryotes. They can remove negative supercoils without leaving nicks in the DNA molecule.
Mechanism:
After the enzyme binds to a DNA molecule and cuts on strand, the free 5' phosphate on the DNA is covalently attached to a tyrosine residue in the enzyme in the case of prokaryotes (the free 3'-phosphate on the DNA is covalently attached in the case of eukaryotes). The DNA strand that has not been cleaved is then passed through the single stranded break. The cleaved strand is then resealed. By this mechanism, the enzyme removes one negative supercoil at a time there by increasing the LK by one in the case of prokaryotes. By forming free 3' or free 5' end DNA- protein covalent intermediates, the free energy of the cleaved phosphodiester bond is preserved so that no energy input is required to reseal the nick.
The type -I topoisomerase from E.Coli acts on negatively supercoiled molecules but not on positively supercoiled molecules. In contrast, type-I topoisomerase from eukaryotic cells can remove both positive and negative supercoils. Type-I topoisomerase reversibly catenates (interlinks) single stranded circles. Topoisomerases-I and Topoisomerases-II are type-I topoisomerases.
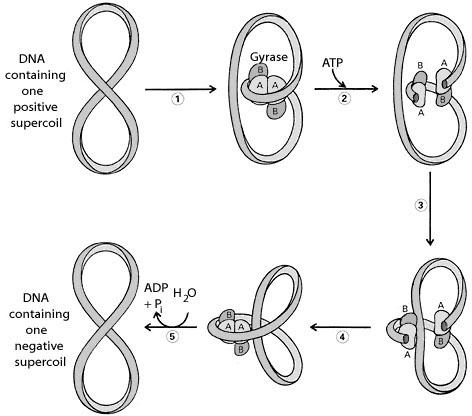
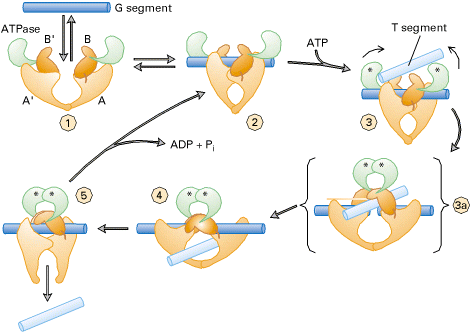
TYPE-II TOPOISOMERASES:
Prokaryotic type-II topoisomerases, which are also known as DNA gyrases, are 375kd proteins that consists of two pairs of subunits designated A and B. DNA gyrase converts a right-handed toroidal supercoil to a left-handed toroidal supercoil. This mechanism is called as sign inversion mechanism. DNA gyrase requires ATP for its function. Hydrolysis of ATP is not required to induce supercoils but is required for the enzyme to turnover and introduces additional supercoils.
The type-II topoisomerase enzymes from mammalian cells cannot increase the superhelicity (super helical density) at the expense of ATP, presumably no such activity is required in eukaryotes since binding of histones increases the potential superhelicity. DNA gyrase has the ability to cut a double stranded DNA molecule, pass another portion of the duplex DNA through the cut, and reseal the cut. It changes the linking number of the DNA by 2.
|
S.No |
Properties |
Prokaryotes |
Eukaryotes |
||
|
Type-I |
Type-II |
Type-I |
Type-II |
||
|
1. |
Molecular Weight |
100,000 |
400,000 |
1,00,000 |
3,09,000 |
|
2. |
Subunits |
Monomer |
Tetramer |
- |
Dimers |
|
3. |
Gene |
Topo - A |
gyr - A gyr - B |
- |
- |
|
4. |
Covalent Intermediate DNA |
At 5'-P |
Subunit A at 5'-P |
At 3'-P |
At 5'- P |
|
5. |
Relaxation |
Mg2+ dependent ATP independent |
ATP dependent |
Mg2+ independent ATP independent |
ATP dependent |
|
6. |
Negative supercoiling |
- |
ATP dependent |
- |
- |
All type-II topoisomerase catalyze catenation and decatenation that is, the linking and unlinking of two different DNA duplexes. Topoisomerase-IV is also a type of type-II topoisomerases. It actively involved in decatenation i.e. termination of DNA replication.
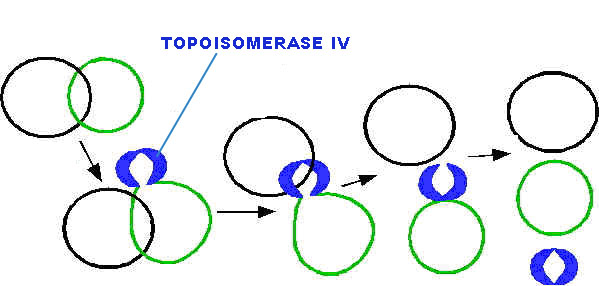
SINGLE STRAND BINDING (SSB) PROTEINS:
Single strand binding protein named because its nature to bind to single strand of DNA which will be formed during open complex formation during initiation of replication. It contains four subunits. The major function of SSB is to prevent recoiling of DNA strands after it’s unwinding by helicases. Thus, SSB plays vital role in replication.