REPLICATION
EVIDENCES FOR THE CONCEPT OF DNA AS THE BEARER OF GENETIC INFORMATION:
The studies that have revealed the chemistry of genes began in Tubingen, Germany in 1869. There, Friedrich Miescher isolated nuclei from pus cells (WBCs) in waste surgical bandages. He found that these nuclei contained a novel phosphorus bearing substance that he named nuclein. Nuclein is mostly chromatin, a complex of DNA and chromosomal proteins. But it took more than seven decades to realize its biological role and substantiate with conclusive experimental evidences to prove that DNA is the bearer of genetic information. Following are the experiments to prove that. They are
a) Griffiths' Experiment (Discovery of Transforming Principle)
b) O.T. Avery, Macleod and McCarty Experiment
c) Hershey and Chase Experiment
d) Transgenic Mice Experiment
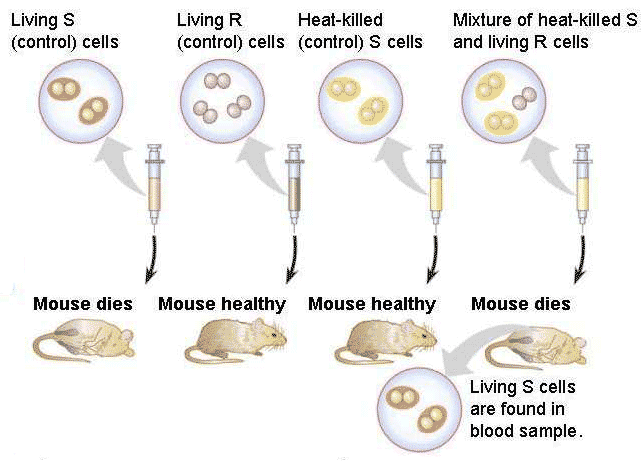
a) FREDRICK GRIFFITHS' EXPERIMENT WITH Pneumococcus: (Discovery of Transforming Principle (1928))
Streptococcus pneumonia (Pneumococcus bacteria) a bacterium that causes pneumonia is of two types.
i) Smooth 'S' type:
It is the virulent form of bacteria. The virulence is due to the presence of capsular gelatinous polysaccharide coating which contains O-antigens through which it recognizes the cells it infects.
ii) Rough 'R' type:
It is a mutant variety and the nonvirulent form of bacteria because it lacks the capsular gelatinous polysaccharide coating.
EXPERIMENT:
Griffith in his experiment with pneumococcus took four groups of mice which were treated differently. To one group, he injected live S-type. To the second group, he injected live R-type. To the third group heat inactivated (i.e. bacteria killed by heat treatment) virulent S-type bacteria and to the fourth group a mixture of heat killed smooth bacteria and live rough bacteria were injected.
OBSERVATION:
The following results were obtained. Neither heat inactivated virulent form nor do the rough forms kill the mice. But the live S-type and mixture of both kills the mice. Post mortem analysis of blood of infected mice of fourth group showed live smooth forms. Moreover, the transformed ones give rise to virulent ones showing that the transformation was permanent.

CONCLUSION:
From the observations it was concluded that the non-virulent (R- form) bacteria got transformed by some components of the dead virulent form which was then called the 'transforming Principle'.
B) ISOLATION AND ESTABLISHING THAT THE TRANSFORMING PRINCIPLE (O.T.AVERY et al., EXPERIMENT - 1944):
O.T.Avery et al., after more than 10 years of intense investigation conclusively proved that the transforming principle was indeed the DNA.
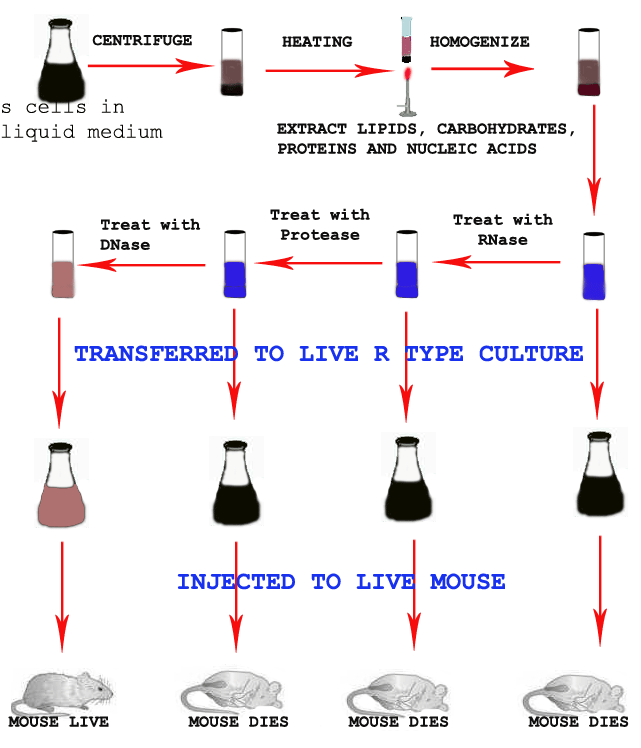
EXPERIMENT:
In their experiments, cell free form of DNA from the virulent form of pneumococcus (S-form) were extracted through the process of centrifugation, heating and homogenization and injected into the colony of non-virulent (R-type). After sometimes, when these colonies were injected into group of mice they found to cause pneumonia.
Similarly the cell free extract, first treated with RNase and a portion mixed with colony of R type and then injected to group of mice. To the remaining RNase treated cell free extract, Protease added and a portion mixed with colony of R type and then injected to group of mice. To the remaining cell free extract treated with RNase and Protease, DNase added. The mixture was then mixed with R-type and injected into another group of mice.
OBSERVATION AND CONCLUSION:
The transforming principle which was identified as DNA transforms the non-virulent bacteria into virulent form of pneumococcus. The results are as follows:
1) Mixture of cell free extract and R-type cause pneumonia and mouse dead.
2) Addition of RNase did not affect the phenomena of transformation which again neglects the possibility of RNA being the transformation factor.
3) Addition of Proteases like Trypsin and Chymotrypsin again did not affect the transformation showing that the transforming principle was not a protein.
4) Addition of DNase (which specifically destroys DNA) to the transforming principle extract abolish transformation, showing that the DNA is the active genetic factor.
Finally, direct physical-chemical analysis also showed the purified transforming substance to be DNA.
The analytical tools, Avery and his colleagues used were the following:
A. ULTRA CENTRIFUGATION:
They spun the transforming substance in an ultracentrifuge to estimate its size. The material with transforming activity was sedimented rapidly, suggesting a very high molecular weight which is characteristic of DNA.
B. ELECTROPHORESIS:
They placed the transforming principle in an electric field to see how rapidly it moved. The transforming activity had a relatively high mobility which is also characteristic of DNA.
C. ULTRAVIOLET ABSORPTION SPECTROSCOPY:
They placed a solution of the transforming substance in a spectrophotometer to see what kind of UV light it absorbed most strongly. Its absorption spectrum matched that of DNA. That is the light it absorbed most strongly had a wavelength of about 260nm, in contrast to protein which absorb maximally at 280nm.
D) ELEMENTARY CHEMICAL ANALYSIS:
This yielded an average nitrogen / phosphorous ratio of 1.67, about what one would expect for DNA, which is rich in both elements, but vastly lower than the value expected for protein, which is rich in nitrogen but poor in phosphorous.
Furthermore Rollin Hotcheleiss had refined and extended Avery's findings. He purified the transforming substances to the point where it contained only 0.02% protein, and showed that it could still change the genetic characteristics of bacterial cells. He went on to show that such highly purified DNA could transfer genetic traits other than R and S.
C) EXPERIMENT OF A.D. HERSHEY AND M.CHASE ON BACTERIOPHAGE T2 (Isotopic studies on bacterial virus T2 - 1952):
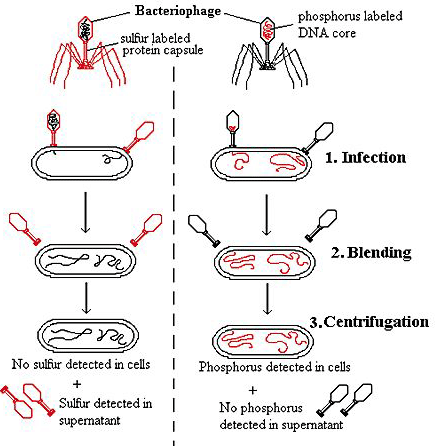
Phage T2 contains genes that allow it to replicate in E.Coli. Since, the phage is composed of DNA and protein only, its genes must be made of one of these substance. To function as genes, one of these substances has to enter into E.Coli. By utilizing this thought experiment performed.
In their experiment, they grew bacteriophage T2 on E.Coli in a medium containing radioactive isotopes 32P and 35S. This labeled the phage capsid which contains no phosphorus with 35S and its DNA which contain no sulphur with 32P.
These labeled phages were added to an unlabelled culture of E.Coli and allowed for sufficient time for the phages to infect the bacterial cells. After this time, this culture was spun for few minutes in a warring blender at 10,000rpm. This treatment subjected the phage infected cells to a very strong sharing force which rupture the connection between viruses and bacteria. The resulting suspension was centrifuged at a speed sufficient to separate bacteria to the bottom of the tube while the phage ghosts (empty phage shells) remained in the supernatant. The centrifuged fractions were then analyzed for the distribution of radioactive substances 32 P and 35S.
RESULTS:
Most of the labeled protein remained in the supernatant which contain 35S. The bacterial fraction (Pellet), which contain most of the 32 P showing that the most of the phage DNA, was in the bacteria. The conclusion was that the genes of this phage are made of DNA.
D) RALPH BRINSTER EXPERIMENT (Transgenic mice producing Experiment - 1982):
Ralph Brinster, in a spectacular demonstration proved that transformation phenomena can also takes place in eukaryotes and DNA being the transforming principle.
He microinjected DNA bearing the gene for rat growth hormone into the nuclei of fertilized mouse eggs and implanted these eggs into the uteri of foster mothers. The resulting 'supermice' which had high levels of rat growth hormones in their serum, grew to nearly twice the weight of their normal litter mates. Such genetically altered animals are said to be transgenic animals. So, in this experiment 'transgenic mice' produced.
From this experiment, it was proved that DNA being the transforming principle in eukaryotes.
RNA AS GENETIC MATERIAL:
In some viruses, RNA is the genetic material. The tobacco mosaic virus that infects tobacco plants consists only of RNA and protein. The single, long RNA molecule is packaged within a rod like structure formed by over two thousand copies of a single protein. No DNA is present in tobacco mosaic virus particles.
In 1955, H. Fraenkel Conrat and R.Williams showed that a virus can be separated invitro into its component parts and reconstituted as a viable virus. This finding led to experiments by Fraenkel Conrat and B. Singer who reconstituted Tobacco Mosaic Virus (TMV) with parts from different strains.

For example, they combined the RNA from the common TMV with the protein from the masked (HR) strain of TMV. They sprayed reconstituted hybrid virus on to tobacco leaf. The tobacco mosaic virus produced during the process of infection was of the type associated with the RNA, not with the protein. Thus it was the nucleic acid (RNA) that was shown to be the genetic material. This was confirmed in subsequent experiments in which pure TMV RNA was rubbed into plant leaves. Normal infection and a new generation of typical, protein coated TMV resulted.
We thus conclude that DNA is the genetic material. In the few viruses that do not have DNA, RNA serves as the genetic material. The only exception to these statements is one type of disease in which transmission is by a protein without accompanying DNA or RNA i.e., in the case of "Prions".
GENE:
The fragment of DNA which can be transcribed into functional RNAs like r-RNA and t-RNA or translated into functional protein is termed as GENE.
CHROMOSOME STRUCTURE:
All types of nucleic acids interact with proteins. Chromosomal DNA forms stable nonspecific complexes with structural proteins that stabilize their tertiary structure; it also forms transient complexes with enzymes and regulatory proteins that modulate DNA and RNA metabolism. Chromosomal Structure of prokaryotes and eukaryotes are discussed below:
GENOME ORGANISATION IN PROKARYOTES (CHROMOSOMAL STRUCTURE IN E.COLI):
Prokaryotes including bacteria contain the genetic material in a fairly compact lump, occupying about one-third of the volume of the cell. Since the genetic material is not compartmentalized and lies naked in the cytoplasm, it is called nucleoid. The nucleoid is a folded structure, containing many supercoiled loops having many independent domains. There are about 100 domains per genome and each domain consists of about 40kb of DNA. The prokaryotic genome is a single replicon.
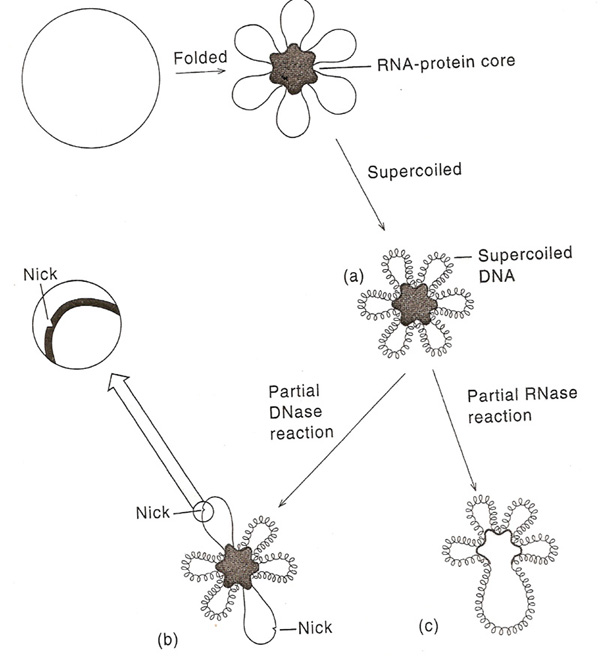
A single chromosome of E.Coli contains about 3x109 Daltons or about 4.5 x 106bp of DNA. If all of this DNA were in a duplex structure stretched end to end, it would be 1.5mm long, which is about 75 cell diameter. But inside the cell, the chromosome is coiled to the size of 2 micrometer. Electron micrographs of E.Coli chromosome suggest a folded circular structure containing 40-100 super coiled loops. This Structure is formed by the following steps i.e. initially circular DNA binds with the RNA-Protein core and forms loops. Secondly, the loops form super coiled structure. The interaction between the loops and RNA - Protein core is not understood. D.E. Pettijohn and his co-workers have provided evidence of such a core. They first showed that the individual supercoiled loops maintained their supercoiling independently of one another. Thus, if a single nick is introduced into one of the loops by limited DNase action. That loops adopts an expanded relaxed conformation, but supercoiling in the other loops is maintained. Limited RNase or protease treatment causes the partial breakdown of the looped structures without interfering with the supercoiling. These results have lead to the conclusion that each of the loops is a domain, the lateral motion of which is restricted by an RNA-Protein core complex.
GENE ORGANIZATION IN PROKARYOTES:
The circular E.Coli chromosome contains enough base pairs to make about 3,000 average-sized genes. The relative positions of over half of these genes are known. The genes are present in a tightly organized form. Coding regions are interspersed with regulatory regions; there is no evidence for significant stretches of DNA with any function. Frequently, genes with a related function are clustered. These clustered genes are usually transcribed into single expression units (messenger RNA) containing the information for the synthesis of several functionally related proteins. Because of this character, in prokaryotes, genes are known as polycistronic in nature. The gene arrangement is explained with the concept of operons like Lac operon etc.
OPERONS:
One characteristic feature of prokaryotic genomes illustrated by E. coli is the presence of operons. An operon is a group of genes that are located adjacent to one another in the genome, with perhaps just one or two nucleotides between the end of one gene and the start of the next. All the genes in an operon are expressed as a single unit. This type of arrangement is common in prokaryotic genomes. A typical E. coli example is the lactose operon (catabolic operon), the first operon to be discovered (Jacob and Monod, 1961), which contains three genes involved in conversion of the disaccharide sugar lactose into its monosaccharide units - glucose and galactose. The monosaccharides are substrates for the energy-generating glycolytic pathway, so the function of the genes in the lactose operon is to convert lactose into a form that can be utilized by E. coli as an energy source. Lactose is not a common component of E. coli's natural environment, so most of the time the operon is not expressed and the enzymes for lactose utilization are not made by the bacterium. When lactose becomes available, it switches on the operon; all three genes are expressed together, resulting in coordinated synthesis of the lactose-utilizing enzymes.
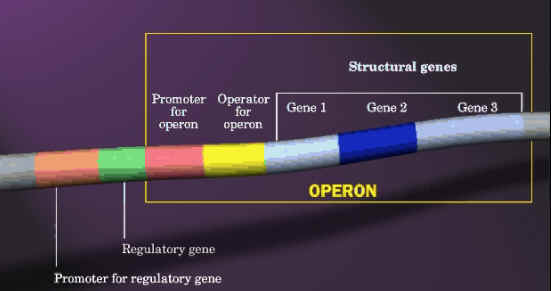
Similarly, there will be another operon namely Trp operon which actively codes for enzymes which are involved in synthesis of tryptophan (anabolic operon). By considering the above two example one might think that operon will contain genes for related metabolism but it is not true. In some cases like the archaeon Methanococcus jannaschii and the bacterium Aquifex aeolicus have operons, but the genes in an individual operon rarely have any biochemical relationship. For example, one of the operons in the A. aeolicus genome contains six linked genes, these genes coding for two proteins involved in DNA recombination, an enzyme used in protein synthesis, a protein required for motility, an enzyme involved in nucleotide synthesis, and an enzyme for lipid synthesis. The main reason for the presence of operons in prokaryotes provides more genes in a small region due to the reduction in space for the presence of regulatory genes for each gene separately.
EUKARYOTIC GENOME ORGANIZATION:
Eukaryotic DNA is contained in a relatively small number of chromosomes which varies according to the species. No direct correlation can be made between the amount of DNA in the nucleus and the number of chromosomes in which it is combined. Somatic cells of each species have two copies or homologous of each chromosome with the exception of the sex chromosome for which the female carries two 'X' chromosomes and the male an 'X' and a 'Y'. Germ cells contain only one copy of each chromosome and it is to these cells that the term haploid chromosome content or haploid DNA content refers. Aneuploid cells have an abnormal chromosome complement which is not necessarily on increase on the diploid condition for each chromosome type so that some may be present in greater numbers than others. The characteristic number and morphology of the chromosome in any particular cell type is known as the karyotype of that cell and is usually determined in metaphase when the chromosome are highly condensed and readily stained by basic dyes. Staining chromosome with basic dyes known as chromosome painting. In interpahase, the chromosomes are spread out in the nucleus and cannot be individually distinguished. Usually chromosome found to have X shape. Chromosome form of genome available only during Mitotic phase (division phase) whereas normally it present in Chromatin form in cells.

The centromere is the region of the chromosome to which spindle fibers attached. The centromere region usually appears to be constricted and the position of this constriction defines the ratio between the length of the two chromosome arms; this ratio is a useful characteristic centromere positions can be categorized as telocentric where centromere present at one end, acrocentric in which centromere is at off center, metacentric in which centromere present at centre and acentric in which chromosome lacks centromere.
Nucleoli are intranuclear organelles that contain rRNA, an important component of ribosomes. Different organisms are differently endowed with nucleoli, which range in number from one to many per chromosome set. The diploid cells of many species have two nucleoli. The nucleoli reside next to secondary constrictions of the chromosomes, called nucleolar organizers, which have highly specific positions in the chromosome set. Nucleolar organizers contain the genes that code for ribosomal RNA.
Chromomeres are beadlike, localized thickenings found along the chromosome during prophase of mitosis and meiosis. Homologous chromosomes tend to have homologous sets of chromomeres. Although they can be useful as markers, their molecular nature is not known.
When chromosomes are treated with chemicals that react with DNA, such as Feulgen stain, distinct regions with different staining characteristics are visually revealed. Densely staining regions are called heterochromatin; poorly staining regions are said to be euchromatin. The distinction refers to the degree of compactness or coiling, of the DNA in the chromosome. Heterochromatin can be either constitutive or Facultative. The constitutive type is a permanent feature of a specific chromosome location. The facultative type is sometimes but not always found at a particular chromosomal location. The pattern of heterochromatin and euchromatin along a chromosome are good cytogenetic markers. Chromatid is one of the two side by side replicas produced by chromosome division. Chromatin is the material of chromosomes, composed of DNA, chromosomal proteins and chromosomal RNA. Each eukaryotic chromosome contains a single, long, folded DNA molecule.
Chromosomes responsible for sexual determination are generally named as allosomes (sex chromosomes) and others named as autosomes (somatic chromosomes). For example, in humans, XX or XY chromosomes considered as allosomes and other 22 pair chromosomes considered as autosomes.
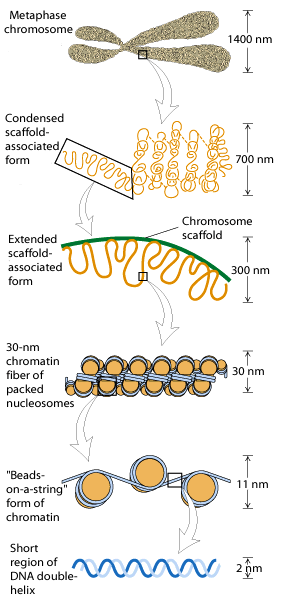
CHROMOSOME PACKING (CHROMATIN ORGANIZATION):
The Progressive levels of chromosome packing are as follows
i) DNA winds onto nucleosome spools
ii) The nucleosome chain coils into a solenoid
iii) The solenoid forms radial loops and the loops attach to a central scaffold
iv) The scaffold plus loops arrange themselves into a giant supercoil.
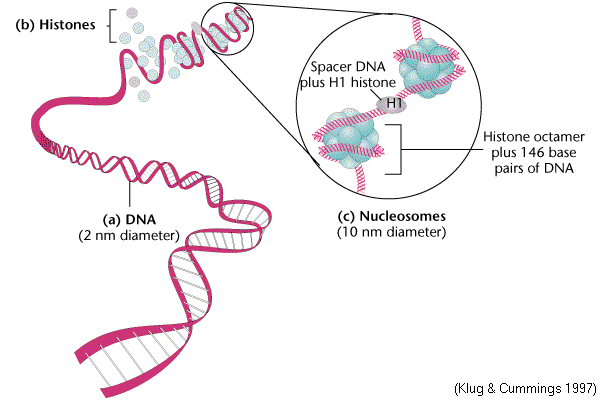
NUCLEOSOME [THE FIRST ORDER OF CHROMATIN FOLDING]:
The first level of chromatin folding was pointed out by Roger Kornberg in 1974. Nucleosome is the basic unit of eukaryotic chromosome. It is a ball of eight histone molecules wrapped about by two coils of DNA. Nucleosomes consists of the octomer Histones [H2A]2, [H2B]2, [H3]2 and [H4]2 in association with approximately 200bps of DNA. The fifth histone, H1, was postulated to be associated in some manner with the outside of the nucleosome. Nucleosome provide the extremely long thin chromosomal fiber with 20A'. The apparent decrease in length of the DNA in the minichromosome is due to the formation of nucleosomes. Infact, the DNA achieves about a seven fold condensation by coiling up into nucleosomes.
Each bead of nucleosome is actually a ball of histones with the DNA wound almost twice around the outside. Each ball contains exactly eight histone molecules, a pair each of H2A, H2B, H3 and H4. A single molecule of histone H1 binds to linker DNA outside the ball and can be removed by more stringent DNase treatment, which digests the linker DNA and yields a nucleosome core particle with about 150bps of DNA.
Nucleosome appears that the core histones are really not arranged in pairs. H3 and H4 form a tetramer at the center of the particle and these tetramers is flanked by H2A-H2B dimers at top left and lower right. Two properties of nucleosomes suggest that their function is fundamentally important. The first is the universality of nucleosomes in eukaryotes. The second is the extreme evolutionary conservation of most of the histones example: H4 from the cow differs only by two aminoacids from that of pea plant.
SOLENOID [SECOND ORDER OF CHROMATIN FOLDING]:
The next thickness, about 30nm (300A'), is due to further winding of the nucleosome to form a hollow coil called a solenoid.
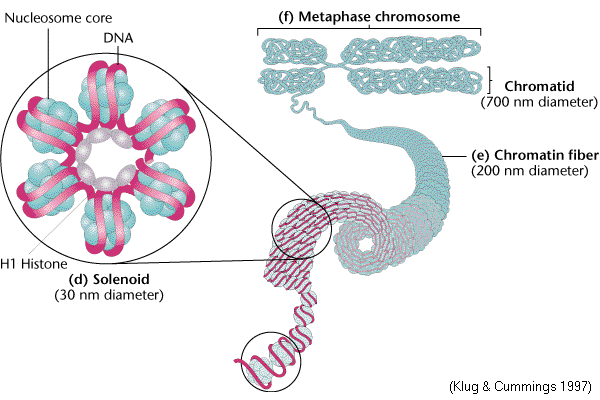
Aaron Klug, who first described the solenoid, traced its formation by making electron micrographs of chromatin in solutions of increasing salt concentration. At very low salt concentration, the chromatin appears as a string of nucleosomes. As the salt concentration rises, coiling takes place, until the typical solenoid structure appear. The DNA achieves another 6 to 7 fold condensation in coiling up in to the solenoid. H1 participates in this coiling because coiling cannot occur in salt treated chromatin that lacks H1. Solenoid structure was stabilized by H1-H1 interactions.
RADIAL LOOPS [THIRD ORDER OF CHROMATIN FOLDING]:
Histone depleted metaphase chromosomes exhibit a central fibrous protein "scaffold" surrounded by an extensive hallow of DNA. The strands of DNA that can be followed are observed to form loops that enter and exit the scaffold at nearly the same point.
It is possible to digest 99% of the DNA from metaphase chromosome preparations leaving behind a morphologically intact central chromosome 'scaffold'. In a similar manner the material which remains when dehistonated interphase chromatin has most of its DNA removed by nuclease treatment is known as the nuclear 'matrix'. The relationship between the scaffold and matrix is unclear.
The scaffold proteins include two high molecular weight proteins of Mw of 1,70,000 and 1,35,000 but the matrix as well as containing an ill defined fibrous protein network is also associated with the nuclear pore lamina complex to which the chromosome are attached.
CHROMATID FORMATION [FOURTH ORDER OF CHROMATIN FOLDING]:
Most of these radial loops have lengths in the range 15 to 30 micrometer, so that when coiled as filaments they would be about 0.7micrometer long. Electron micrograph of a metaphase chromosome strongly suggests that the chromatid fibers of metaphase chromosomes are radially arranged. If the observed loops correspond to these radial fibers. They each contribute 0.3micrometer to the diameter of the chromosome. Taking into account the 0.4micron width of the scaffold, this model predicts the diameter of the chromatid of the metaphase chromosome to be 1 micron. Therefore, the total diameter of the chromosome will be around 1.4micron to 2 micron. A typical human chromosome, which contains ~ 140million bps, would therefore have 2000 of these 70kb radial loops.
EUKARYOTIC GENE ORGANIZATION:
According to the classical definition, a gene is a unique component of the genome occupying a particular locus. Each gene codes for a protein product and can be identified by mutations that impede the protein function. Such genes are structural genes. Another class of genes is found in DNA that is in the form of multiple sequences. These genes code for the same protein or closely related proteins. For the purpose of identifying and characterizing structural genes, mRNA provides the ideal intermediate and it can be subsequently used to translate into protein it codes for. It may be noted that most structural genes lie in non-repetitive DNA. Structural genes may have three types of organization:
a. They may consist of a DNA sequence that code for a particular protein.
b. Besides structural genes, there could be other sequences in the genome that code for related proteins repetitive and moderately repetitive.
c. Structural genes may be repeated, i.e. there may be more than one copy of DNA sequence coding for a particular protein.
The eukaryotic individual genes can be interrupted, there can be multiple and identical copies of particular sequences and there may be large sequences of nucleotides that do not encode for protein. Another important feature of eukaryotes is the compartmentalization of the nucleus and the cytoplasm that may create differences in the gene expression.
The eukaryotic gene structure is as follows:
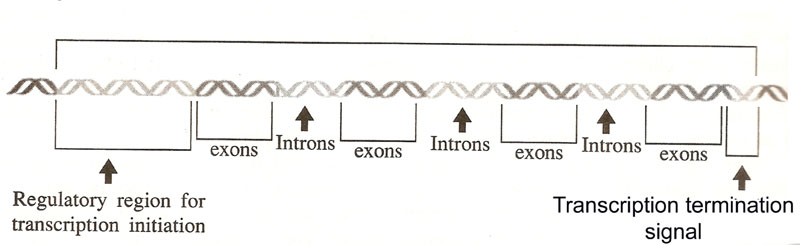
The sequences of the DNA comprising the gene are thus divided into two categories. The exons comprise of the regions that are represented in the mRNA and used to produce the protein product. The introns are missing from the mRNA. The process of gene expressions involves a new step, one that does not occur in bacteria. The DNA produces a copy of RNA with the exact copy of gene sequence. This RNA is only a precursor and in order to produce a functional mRNA, it undergoes splicing to remove introns (non-coding sequences).
In eukaryotes, the relationship between gene and its RNA product is to be understood and it is summarized as follows:
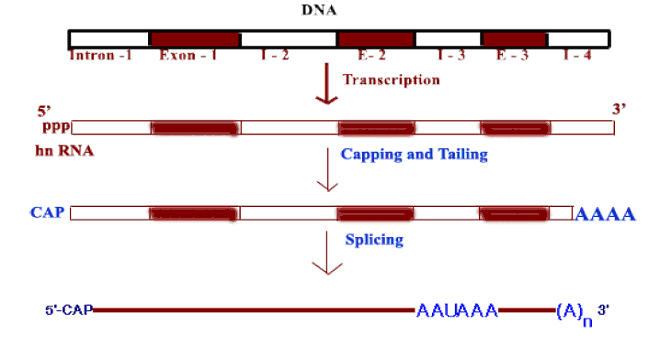
An interrupted gene consists of alternating series of exons and introns. Exon sequences are present in the RNA. Introns are intervening sequences that are removed from the primary transcript and are absent from the mature mRNA. A gene must start and end with exons, corresponding to 5 and 3 ends of RNA.
An intervening sequence is an invariant feature. The intervening sequences vary enormously in both number and size, but all classes of genes may be interrupted. The sequences of interrupted genes are same as in case of mature RNA product. Thus genes split rather than be dispersed. Introns of nuclear genes generally have termination codons in all reading frames and an interrupted gene retains the same structure in all tissues. All intervening sequences must be removed before they are translated. The length of the intron varies widely. They may be as short as 14 bp or as long as 46 bp in different genes. Introns are non-repetitive and at the exon-intron junctions, consensus sequences are located.
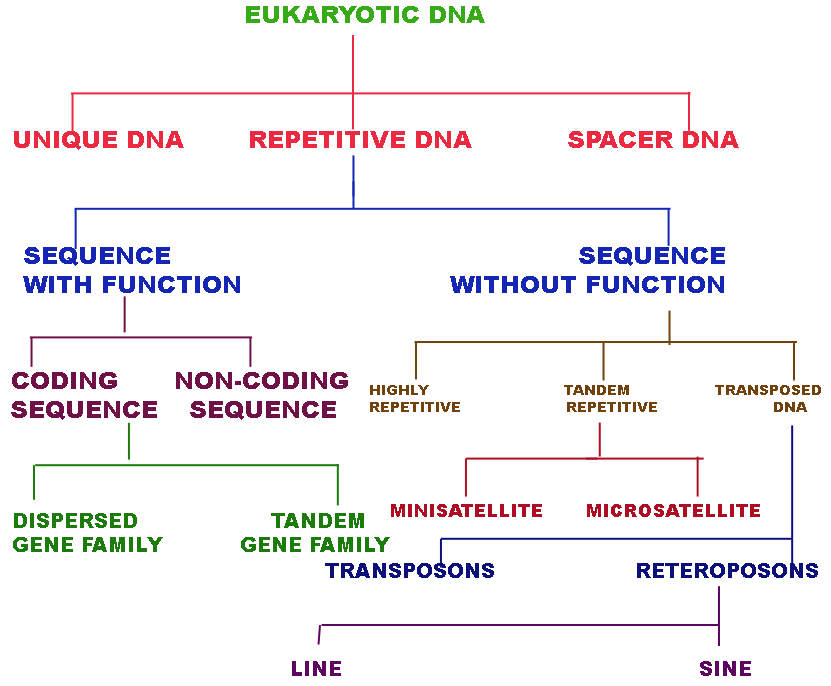
EUKARYOTIC DNA ORGANIZATION:
Eukaryotic DNA found to consists of mainly three parts namely Single copy functional gene, Repetitive DNA and Spacer or Selfish DNA. Sequences that are present in more than one copy in each genome are described as repetitive DNA and the number of copies present per genome is called the repetition frequency (f), which is defined by the following ratio:
Repetition frequency (f) = Chemical Complexity / Kinetic Complexity
Most of the functional genes of eukaryotes present in single gene form.
UNIQUE DNA:
DNA which was available in single copy i.e a length of DNA with no repetitive nucleotide sequence referred as unique DNA. The unique DNA makes up the structural genes much of it is transcribed. They are functional genes.
REPETITIVE DNA:
In Repetitive DNA, there are two types are found to present namely functional sequences and nonfunctional sequences.
SEQUENCES WITH FUNCTION:
Functional sequences further classified into two groups namely coding and non coding sequences. Coding sequences are further divided into two classes namely Dispersed gene families and Tandem gene families.
Families of homologous genes spread throughout the genome codes several types of proteins. These gene families are referred as Dispersed gene families. For example: histones contain genes from 100 to 1000 and actins from 5 to 30. Some genes within families have become nonfunctional, giving rise to untranscribed pseudogenes.
If genes of particular type protein present tandemly (adjacently) present then they are said to be tandem gene family arrays. Genes for the proteins which are needed in large amounts by cell are usually present in tandem array. For example: human tRNA contains 50 chromosomal sites and each site contains between 10 and 100 copies.
There are certain sequences which are found to be present in tandem arrays and they are not either transcribed into RNA or translated to protein. But they found to have definite function are referred as noncoding functional sequences. For example: telomeres in chromosomes. In human it is TTAGGG, this is used to replicate the end region of the linear DNA molecule i.e. the gap created by the primer at 5' end during replication.
SEQUENCES WITHOUT FUNCTION:
Repetitive sequences without function further divided into three groups namely Highly repetitive DNA, Tandem Repetitive and Transposed Sequences.
HIGHLY REPETITIVE DNA:
Refer Cot curve Notes.
TANDEM REPETITIVE DNA:
Tandem repetitive DNA further classified into minisatellite and microsatellite DNA.
MINISATELLITE DNA (VARIABLE NUMBER OF TANDEM RPEAT [VNTR]):
A special class of tandem repeat shows variable number at different chromosomal positions and in different individual members of a species. This type of repeat is called as Variable Number of tandem repeat or minisatellite DNA. The VNTR loci in humans are 1 to 5kb sequences consisting of variable numbers of a repeating unit from 15 to 100 nucleotides long. They are used extensively in forensic medicine through DNA fingerprinting technique.
MICROSATELLITE DNA:
It is the class of dispersed repetitive DNA which is composed of dinucleotide repeats. Because the number of repeats varies between individuals, this type of DNA has been most useful in providing a dense array of molecular markers for mapping the human genome. The most common type consists of repeats of CA and its complement GT.
TRANSPOSED SEQUENCES:
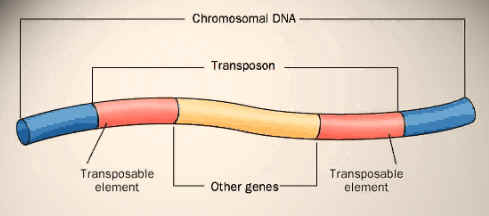
A large proportion of a eukaryotic genome is composed of repetitive elements that have propagated within the genome by making copies of them, which can move into new locations. These elements are collectively called transposed sequences. Those elements that move as DNA are called as transposans. Many genomes have multiple copies of such elements or truncated versions of them dispersed throughout the genome. Tn3 is one of the examples for DNA transposons. DNA sequences that propagate themselves through the action of reverse transcriptase, enzyme that can make a DNA strand from RNA referred as retrotransposons. There are two subtypes are present namely LINEs and SINEs.
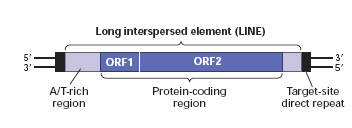
LONG INTERSPERSED ELEMENTS [LINEs]:
Sequences of DNA upto seven thousand bps in length interspersed in eukaryotes in many copies are referred as LINEs. Their function is unknown. They are type of large repetitive DNA segments found throughout the genome. For example: Ty elements of Yeast.
SHORT INTERSPERSED ELEMENTS [SINEs]:
Sequences of DNA interspersed in eukaryotic chromosome in many copies are referred as SINEs. Their function is unknown. They are found throughout the eukaryotic genome. Alu sequence is an example for SINEs.
SPACER DNA:
The final category of eukaryotic DNA type is Spacer DNA. DNA found between genes whose function is only to provide space probably and it is basically what is left after all recognizable units have been ignited. Little is known about spacer DNA.
SELFISH DNA:
A segment of the genome with no apparent function although it can control its own copy number is referred as selfish DNA. It is s type of DNA that exists only for the purpose of existing and is never exposed to other rigors of phenotype.
Cot CURVES or Cot PLOTS:
When DNA is heated, it denatures or unwinds into single strands. When it is cooled, it renatures. The rate of renaturation depends on the concentration of nucleotide strands and their sequences when the temperature kept constant and the sample is broken into small uniform pieces. This forms the principle for cot curve which was proposed by R. Britten.
If 'Co' is the original concentration of single stranded DNA in moles per liter and t is elapsed time in seconds, then their products provides a scale of renaturation called Cot or Cot. If Co kept constant, then Cot depends upon time scale. When Cot values are plotted against the fraction of DNA renatured, the curve is called as Cot curve or Cot plot. The midpoint, referred to as Cot1/2 value estimates the amount of homology within the DNA or length of unique DNA i.e. length of DNA in a sample that has no repeated sequences. The following Cot curve shows same shape to different DNAs but their location differs from eachother. The right hand side shift of the curve indicates that the more slowly the single stranded nucleic acids reassociate and also increase in the length of unique sequence.
Cot curve for prokaryotic DNA is as follows:
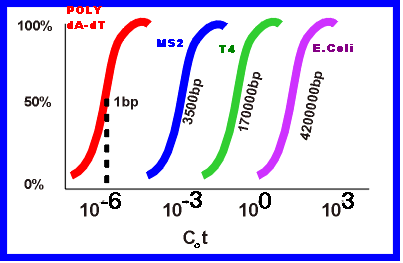
Cot curve for eukaryotic DNA is as follows:
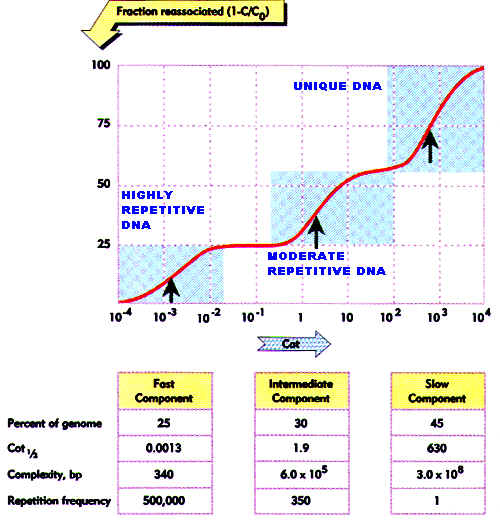
It is actually made up of three separate curves. On the basis of Cot1/2 values and the proportion of the genome that each segment comprises, the degree of repetitiveness of each segment can be determined. There is a highly repetitive segment, a segment of intermediate repetitiveness and a segment of unique DNA. These segments make up of about 25%, 30% and 45% respectively of the total eukaryotic DNA.
C Value Paradox represents morphological complexicities of an organism. It depends upon the amount of DNA and C value.
DNA REPETITION IN EUKARYOTES:
Repetitiveness of DNA is studied by using DNA-DNA hybridization and Cot curves. Depending upon the repetitive nature, there are three classes of DNAs namely Highly repetitive DNA, Intermediate repetitive DNA and Unique DNA.
HIGHLY REPETITIVE DNA:
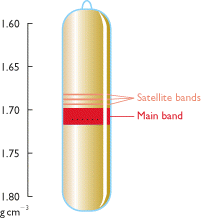
It presents greater than 106 copies per haploid genome. Satellite DNA is the example for highly repetitive DNA when genomic DNA is spun for a long time in a ceisum chloride density gradient in an ultracentrifuge, the DNA settles into one prominent visible band. However, few lighter bands are often visible, distinct from the main DNA band. DNA in such bands is referred as Satellite DNA. Such satellite DNA consists of multiple repeats of short nucleotide sequences, stretching to as much as hundreds of kilobases in length. There can be either one or several basic repeating units, but usually they are less than 10 bases long. There are three satellites DNA in Drosophila Virilis.
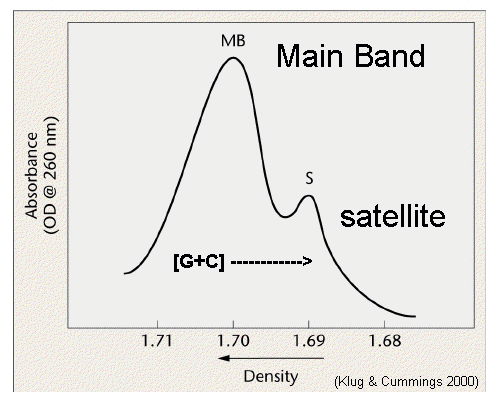
Blocks of satellite sequences are localized to regions around the centromeres of metaphase chromosomes by a technique called Insitu hybridization. In this technique, radioactively labeled RNA copies of the satellite sequence made invitro are hybridized to squashed metaphase cells after they have been treated with alkali to denature the chromosomal DNA. Subsequently the position of the satellite sequences can be visualized by autoradiography.
Because the centromeric repeats are a nonrepresentative sample of the genomic DNA, the G+C content can be significantly different from the rest of the DNA. For this reason, the DNA forms a separate satellite band in an ultracentrifuge density gradient. There is no demonstrable function for centromeric repetitive DNA, nor is there any understanding of its relation to heterochromatin or to genes in the heterochromatin. some organisms have staggering amounts of this DNA, for example, as much as 50% of Kangaroo DNA can be centromeric satellite DNA.
INTERMEDIATELY REPETITIVE DNA:
Intermediately or moderately repetitive DNA in eukaryotes occurs in at least three categories in the genome namely
i) Dispersed probably nontranscribed DNA such as Alu family
ii) Transcribed genes in many copies that are virtually identical such as ribosomal RNA and histone genes.
iii) Trnascribed genes in many copies that have diverged from each other such as antibody, collagen and globin gene.
Moderately repetitive DNA was present less than 106 times. In many mammals, a large portion of the intermediately repetitive DNA is composed of many copies of a short sequences dispersed throughout the genome. For example, in humans, there may be about 3,00,000 copies of a 300bp sequence. Because this sequence is cleaved by the restriction endonuclease Alu-I , it is called Alu family. The exact role of this DNA is not clear at the moment. The possibility exists that some of the members of this family are transcribed into small 7S RNAs. In Some cases, larger RNAs contain Alu sequences. Alu may be involved in the numerous origins of DNA replication along eukaryotic chromosomes or it may be involved in creating secondary structure in messenger RNAs.
Several types of genes create a product that is needed in such large quantity that one copy of the gene could not fulfill the cell's needs. So, that they exists in multiple copies. For example: rRNA genes. Human beings have about 200 copies of the major ribosomal RNA genes and about 2000 copies of 5Sr-RNA genes. Fruit flies have about 200 and 100 copies of the two of multiple copies of a gene which is still not enough. In such cases, the cell must then resort to gene amplification to increase the number of gene copies. For example during Oogenesis, rRNA genes amplified.
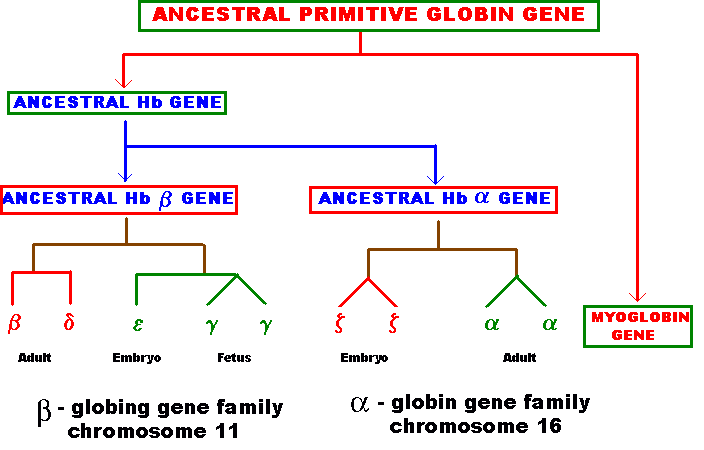
Several types of genes occur in similar but not identical forms. These forms produced by the process of gene duplication. The copies diverged in function. These gene families include globin genes, Ig genes, chorion protein genes in insect egg shell and Drosophila heat shock genes. For example primitive globin gene is initially converted into two globin genes by gene duplication process namely hemoglobin gene and myoglobin gene. These two gene sequences found to be similar but not identical. Because of this, these genes have similar function. Similarly hemoglobin gene undergoes further duplication to form types like Alpha, Beta, Gamma, Delta, Epsilon and Zeta. Eventhough hemoglobin with different combination of polypeptides has oxygen transport function; they differ in their affinity to oxygen. This is the reason for the existence of different polypeptide chain types.
UNIQUE DNA:
DNA, which available in single copy i.e a length of DNA with no repetitive nucleotide sequence was referred as unique DNA. The unique DNA makes up the structural genes much of it is transcribed. They are functional genes.
PSEUDOGENES:
Eukaryotic genes may not be always alone. DNA hybridization analysis has revealed that when genomic fragments are isolated, other sequences are also present that are related to their exons. These additional sequences are sometimes functional genes or at times may be pseudogenes, which are rendered non-functional, but were functional once upon a time. Their inactive nature may be due to accumulation of many interactions over a period of time.
Pseudogenes consist of sequences that are related to functional genes, but cannot be translated into functional proteins. There are some pseudogenes that have the same structure as the functional genes that correspond to exons and introns in the same locations. It has been suggested that these genes are rendered inactive due to mutations, hence cannot be expressed. The mutations are generally responsible for removing the signal sequences initializing transcription, for preventing splicing, or terminating transcription prematurely. This means that pseudogenes are caused by a number of deleterious mutations in the functional gene over a period of time. Pseudogenes are formed in a number of systems that include globin, immunoglobins and histocompatibility antigens.