Basic Concepts of Protein
Structure
|
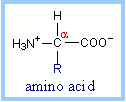
Proteins are polymers of amino
acids. Each amino acid has the same fundamental structure, differing only in
the side-chain, designated the R-group. The carbon atom to which the amino
group, carboxyl group, and side chain (R-group) are attached is the alpha
carbon (Ca).
 |
|
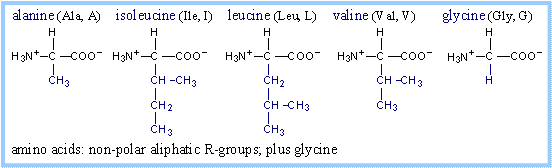
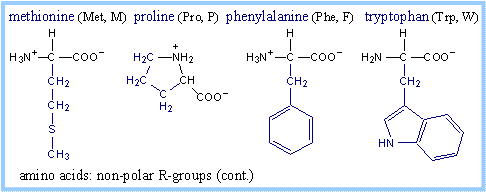
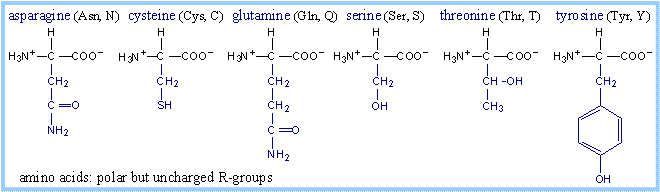
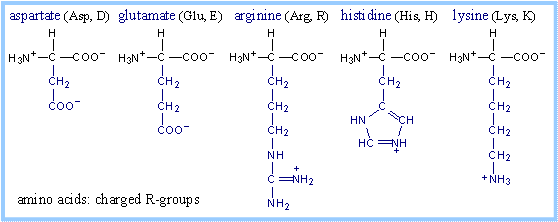
Structures of amino acids, with their
R-groups in blue, are shown below with their three-letter and one-letter
abbreviated names. Glycine has just a hydrogen atom in place of an R-group. At
physiological pH, some amino acid R-groups are charged, because of dissociation
or association of a proton by, e.g., a carboxyl or amino group. Note that
histidine is shown charged, but its pK is close to the physiological range. Some
side-chain groups that are uncharged at the near-neutral pH of the cytosol or
extracellular space, may dissociate or gain a proton in the microenvironment of
an enzyme active site.
Protein structure is studied under
four different levels namely Primary structure, Secondary structure, Tertiary
structure and Quaternary structure.
|
Protein Primary Structure :
Peptide bonds link amino
acid residues within proteins. The sequence of covalently linked amino acids
is known as the primary structure of a protein.
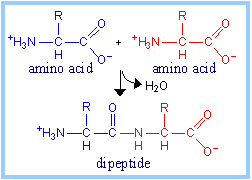
The reaction by which two
amino acids are joined in a peptide bond, with elimination of H2O,
is represented at right. A protein is a polypeptide, a linear polymer of
many amino acids, linked by peptide bonds.
|
|
|
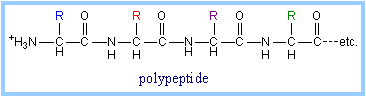
The peptide linkages, along with the
a-carbon atoms to which R-groups
are attached, form the protein backbone, with sequence NCCNCCNCCNCC...
|
|
|
Protein
Secondary Structure:
Segments of polypeptides often
fold locally into stable structures that include
a-helices and
b-pleated sheets.
a-Helix.
In an a-helix (alpha helix), the
polypeptide backbone follows a helical path. There are 3.6 amino acid
residues per turn of the helix. Some protein domains assume other helical
structures, but the a-helix is most
common.
 |
|
|
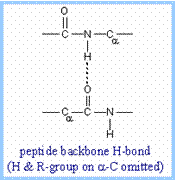
An a-helix
is stabilized by hydrogen bonds between backbone amino and carbonyl groups
and those in the next turn of the helix, represented as N-H····O=C.
The hydrogen and oxygen atoms are attracted to one another because the H
atom carries a partial positive charge and the O atom carries a partial
negative charge, due to unequal sharing of electrons in N-H and O=C bonds.
|
|
|
In an a-helix,
the amino acid R-groups protrude out from the helically coiled polypeptide
backbone. The surface of an a-helix
largely consists of the R-groups of amino acid residues. |
|
|
Some amino acids have a
greater tendency to be found within an a-helix.
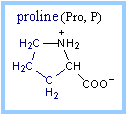
The amino acid proline tends to interrupt an
a-helix. Its fused ring, which
includes the a-carbon and the
peptide-forming amino N, prevents the polypeptide backbone from assuming a
conformation compatible with an a-helix
in the vicinity of a proline.
 |
|
Beta pleated sheets:
Another common secondary
structure is the b sheet (beta sheet).
In a b sheet, strands of protein lie
adjacent to one another, interacting laterally via H bonds between backbone
carbonyl oxygen and amino H atoms. The strands may be parallel (N-termini of
both strands at the same end) or antiparallel.
Because of the tetrahedral nature of carbon
bonds, a b-sheet is puckered, leading
to the designation pleated sheet.
R groups of amino acids in a
b-strand alternately point to one side
or the other of a b-strand. Hence every
other amino acid is exposed on one side or the other of a
b-sheet.
|
A common structural motif,
the b-barrel, is equivalent to a
b-sheet that is rolled up to form a
cylinder. An example of a protein that consists mainly of a
b-barrel is the bacterial channel
porin.

In some cases, a
b-barrel may be partly flattened,
to form what is called a b-clam
structure. |
|
|
Other common folding motifs
involve combinations of a-helices
and b-strands. One example is the
a,b-barrel,
explored elsewhere in an exercise on
Triose Phosphate
Isomerase (TIM).

|
|
Tertiary
protein structure:
It refers to the complete three dimensional folding of a protein. Stabilization
of a protein's tertiary structure may involve interactions between amino acids
located far apart along the primary sequence. These may include:
- weak interactions such as
hydrogen bonds and Van der Waals interactions.
- ionic bonds involving
negatively charged and positively charged amino acid side-chain groups.
- disulfide bonds, covalent
linkages that may form as the thiol groups of two cysteine residues are
oxidized to a disulfide: 2
R-SH
® R-S-S-R.
Interactions with the aqueous solvent, known as
the hydrophobic effect results in residues with non-polar side-chains typically
being buried in the interior of a protein. Conversely, polar amino acid
side-chains tend to on the surface of a protein where they are exposed to the
aqueous milieu. There are, however, many exceptions in which polar residues are
buried or non-polar residues exposed on the surface of a protein. Such atypical
locations might be stabilized, e.g., by interaction of amino acid side-chains
with enzyme prosthetic groups or other ligands, by interactions between amino
acid side-chains, or by association of proteins with lipid
membranes,
etc.
|
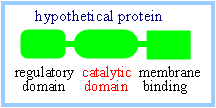
Many
proteins have a modular design with multiple distinct domains resulting from
gene fusion events during evolution. A domain with a particular structure
and function may be part of the structure of several otherwise distinct
proteins. For example, an enzyme's primary structure may include a segment
that folds to produce an active site with particular catalytic activity,
plus other segments that may mediate regulation of the enzyme or binding of
the enzyme to a membrane.
 |
|
|
Quaternary
protein structure :
It refers to
the regular association of two or more polypeptide chains to form a complex.
A multi-subunit protein may be composed of two or more identical
polypeptides, or it may include different polypeptides. Quaternary structure
tends to be stabilized mainly by weak interactions between residues exposed
on surfaces polypeptides within a complex.

A multimeric complex may be important to
enzyme activity, e.g., in cases in which an active site is formed by
residues from more than one polypeptide subunit, or when adjacent active
sites may be involved sequentially in catalysis of a complex reaction.
Regulation of functional activity may involve cooperative interactions among
protein subunits in a complex. |
|













