PHOTOSYNTHESIS
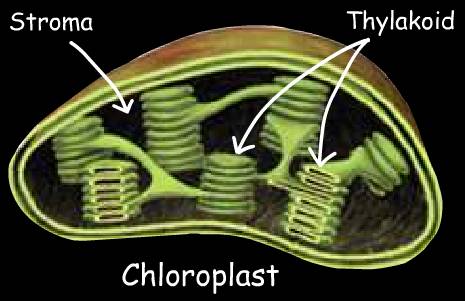
In plants, the light reactions of photosynthesis occur in the thylakoid membrane of the chloroplast. They involve the photosynthetic reaction centers known as Photosystem I and Photosystem II. Both of them are transmembrane protein complexes that each contain a variety of redox-active prosthetic groups. Photosystem I is responsible for reducing NADP+ to NADPH; Photosystem II oxidizes water to diatomic oxygen.

PIGMENTS:
A pigment is any substance that absorbs light. The color of the pigment comes from the wavelengths of light reflected (in other words, those not absorbed). Chlorophyll, the green pigment common to all photosynthetic cells, absorbs all wavelengths of visible light except green, which it reflects to be detected by our eyes. Black pigments absorb all of the wavelengths that strike them. White pigments/lighter colors reflect all or almost all of the energy striking them. Pigments have their own characteristic absorption spectra, the absorption pattern of a given pigment.
Chlorophyll is a complex molecule. Several modifications of chlorophyll occur among plants and other photosynthetic organisms. All photosynthetic organisms (plants, certain protistans, prochlorobacteria, and cyanobacteria) have chlorophyll a. Accessory pigments absorb energy that chlorophyll a does not absorb. Accessory pigments include chlorophyll b (also c, d, and e in algae and protistans), xanthophylls, and carotenoids (such as beta-carotene). Chlorophyll a absorbs its energy from the Violet-Blue and Reddish orange-Red wavelengths, and little from the intermediate (Green-Yellow-Orange) wavelengths.
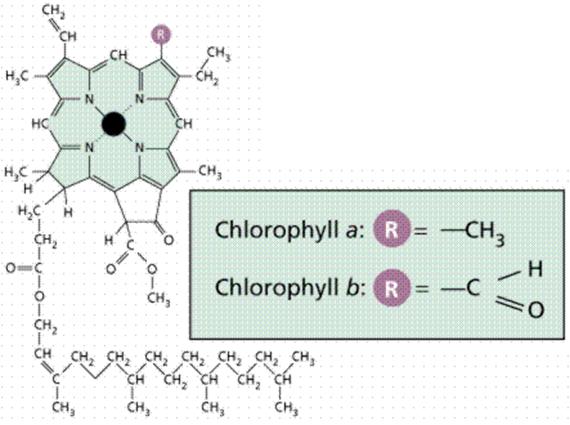
Carotenoids and chlorophyll b absorb some of the energy in the green wavelength. Why not so much in the orange and yellow wavelengths? Both chlorophylls also absorb in the orange-red end of the spectrum (with longer wavelengths and lower energy). The origins of photosynthetic organisms in the sea may account for this. Shorter wavelengths (with more energy) do not penetrate much below 5 meters deep in sea water. The ability to absorb some energy from the longer (hence more penetrating) wavelengths might have been an advantage to early photosynthetic algae that were not able to be in the upper (photic) zone of the sea all the time.
PROCESS:
Photosynthesis is a two stage process. The first process is the Light Dependent Process (Light Reactions), requires the direct energy of light to make energy carrier molecules that are used in the second process.
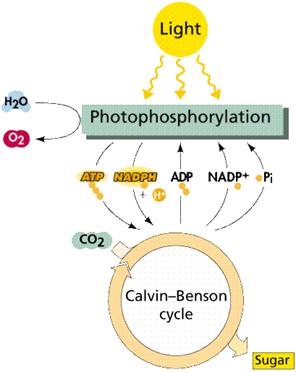
The Light Independent Process (or Dark Reactions) occurs when the products of the Light Reaction are used to form C-C covalent bonds of carbohydrates. The Dark Reactions can usually occur in the dark, if the energy carriers from the light process are present. Recent evidence suggests that a major enzyme of the Dark Reaction is indirectly stimulated by light, thus the term Dark Reaction is somewhat of a misnomer. The Light Reactions occur in the grana and the Dark Reactions take place in the stroma of the chloroplasts.
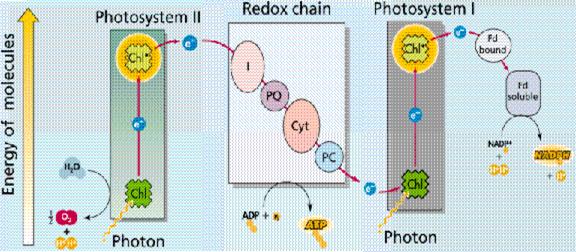
In the Light Dependent Processes (Light Reactions) light strikes chlorophyll a in such a way as to excite electrons to a higher energy state. In a series of reactions the energy is converted (along an electron transport process) into ATP and NADPH. Water is split in the process, releasing oxygen as a by-product of the reaction. The ATP and NADPH are used to make C-C bonds in the Light Independent Process (Dark Reactions).
In the Light Independent Process, carbon dioxide from the atmosphere (or water for aquatic/marine organisms) is captured and modified by the addition of Hydrogen to form carbohydrates (general formula of carbohydrates is [CH2O]n). The incorporation of carbon dioxide into organic compounds is known as carbon fixation. The energy for this comes from the first phase of the photosynthetic process. Living systems cannot directly utilize light energy, but can, through a complicated series of reactions, convert it into C-C bond energy that can be released by glycolysis and other metabolic processes.
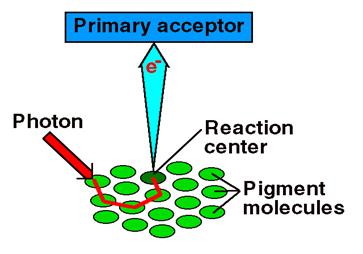
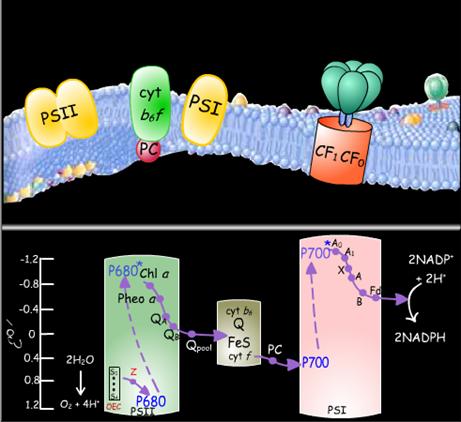
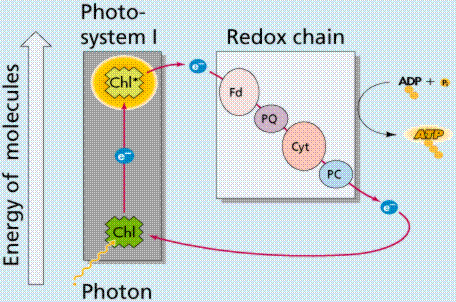
Photosystems are arrangements of chlorophyll and other pigments packed into thylakoids. Many Prokaryotes have only one photosystem, Photosystem II (so numbered because, while it was most likely the first to evolve, it was the second one discovered). Eukaryotes have Photosystem II plus Photosystem I. Photosystem I uses chlorophyll a, in the form referred to as P700. Photosystem II uses a form of chlorophyll a known as P680. Both "active" forms of chlorophyll a function in photosynthesis due to their association with proteins in the thylakoid membrane.
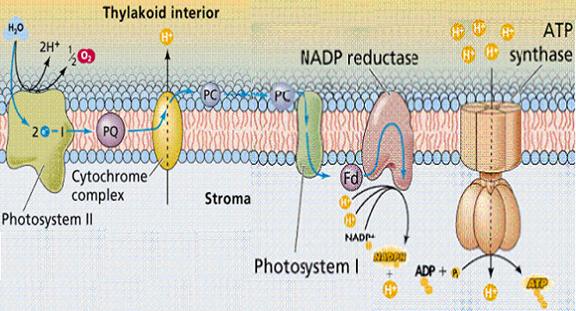
Photophosphorylation is the process of converting energy from a light-excited electron into the pyrophosphate bond of an ADP molecule. This occurs when the electrons from water are excited by the light in the presence of P680. The energy transfer is similar to the chemiosmotic electron transport occurring in the mitochondria. Light energy causes the removal of an electron from a molecule of P680 that is part of Photosystem II. The P680 requires an electron, which is taken from a water molecule, breaking the water into H+ ions and O-2 ions. These O-2 ions combine to form the diatomic O2 that is released. The electron is "boosted" to a higher energy state and attached to a primary electron acceptor, which begins a series of redox reactions, passing the electron through a series of electron carriers, eventually attaching it to a molecule in Photosystem I. Light acts on a molecule of P700 in Photosystem I, causing an electron to be "boosted" to a still higher potential. The electron is attached to a different primary electron acceptor (that is a different molecule from the one associated with Photosystem II). The electron is passed again through a series of redox reactions, eventually being attached to NADP+ and H+ to form NADPH, an energy carrier needed in the Light Independent Reaction. The electron from Photosystem II replaces the excited electron in the P700 molecule. There is thus a continuous flow of electrons from water to NADPH. This energy is used in Carbon Fixation. Cyclic Electron Flow occurs in some eukaryotes and primitive photosynthetic bacteria. No NADPH is produced, only ATP. This occurs when cells may require additional ATP, or when there is no NADP+ to reduce to NADPH. In Photosystem II, the pumping to H ions into the thylakoid and the conversion of ADP + P into ATP is driven by electron gradients established in the thylakoid membrane.
NON-CYCLIC PHOTOPHOSPHORYLATION:
P680, the photon-absorbing center of Photosystem II, consists of one or possibly two closely associated molecules of chlorophyll a. The absorption of a photon of light puts P680 into an excited state known as P680*. This excitation induces P680* to rapidly donate an electron to a nearby molecule of pheophytin a, a process that appears to be influenced by an intervening molecule of chlorophyll a. The resulting cation, P680+, eventually regains its electron by abstracting it from water through the intermediacy of a tyrosine radical named Z and the oxygen-evolving complex, which contains four manganese ions. It requires four consecutive light-induced electron transfers for PSII to convert two molecules of water to one molecule of diatomic oxygen.
Photosystem II binds two molecules of plastoquinone, which chemically resembles ubiquinone in the mitochondrial electron transport chain. The electron received by pheophytin a is rapidly transferred to the plastoquinone molecule designated QA and then is transferred to the second plastoquinone molecule which is called QB, to yield the radical anion QB-. A second photoexcitation of P680 subsequently induces the transfer of a second electron to QB- to yield QB2-, the plastoquinol dianion. QB2- then picks up two protons at the stromal surface of the thylakoid membrane to form QBH2, which is released into the thylakoid membrane and is replaced by a plastoquinone molecule from the pool of plastoquinones in the membrane. PSII is thereby returned to its initial state ready to mediate the photoreduction of the newly bound plastoquinone molecule.
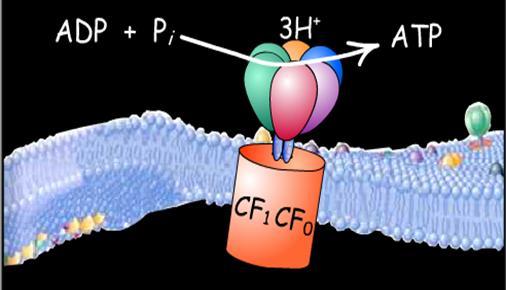
Plastoquinol molecules from the membrane-bound pool bind to the cytochrome b6f complex. This complex contains one subunit each of cytochrome f, cytochrome b6, and an iron-sulfur protein. Two electrons from a plastoquinol molecule pass through the complex one at a time, via a Q cycle, much as occurs in the reduction of the mitochondrial cytochrome bc1 complex by ubiquinol. In the process, four protons are translocated across the thylakoid membrane from the stroma to the thylakoid lumen. The dissipation of the proton gradient that is generated in this manner drives the synthesis of ATP by the CF1CF0-ATPase, which closely resembles the mitochondrial F1F0-ATPase.
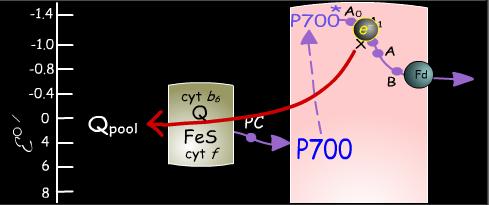
Electrons are transmitted, one at a time, from the cytochrome b6f complex to Photosystem I by the mobile electron carrier plastocyanin. Plastocyanin is a peripheral membrane protein whose bound copper ion is reversibly oxidized from its Cu(I) state to its Cu(II) state. Thus, plastocyanin functions similarly to cytochrome c in the mitochondrial electron transport chain.
The photon absorbing center of Photosystem I is designated P700. In a manner analogous to P680 in Photosystem II, P700 absorbs a photon, resulting in excitation of an electron that it donates to a chain of electron carriers in the complex. Photo-oxidized P700+ then accepts an electron from plastocyanin to regenerate P700.
Returning to the electron lost from the excited P700*, this electron can follow one of two pathways. The more frequently used route is the noncyclic pathway in which electrons are transferred from Photosystem I to the soluble, iron-sulfur-containing protein ferredoxin [Fd]. Two ferredoxin molecules, resulting from two consecutive excitations of Photosystem I, each transfer one electron to the enzyme ferredoxin-NADP+ reductase, which in turn uses the two electrons to reduce NADP+ to NADPH. NADPH is the noncyclic pathway's final product.
CYCLIC PHOTOPHOSPHORYLATION:
The other, less common, pathway for electrons traveling from Photosystem I is the cyclic pathway. Electrons travel from Photosystem I through cytochrome b6 to the plastoquinone pool. In this process, protons are translocated across the thylakoid membrane to increase the proton gradient whose dissipation drives ATP synthesis. The cyclic pathway does not generate NADP+. Moreover, because it does not require the action of Photosystem II, it does not result in the generation of diatomic oxygen.
Halobacteria, which grow in extremely salty water, are facultative aerobes, they can grow when oxygen is absent. Purple pigments, known as retinal (a pigment also found in the human eye) act similar to chlorophyll. The complex of retinal and membrane proteins is known as bacteriorhodopsin, which generates electrons which establish a proton gradient that powers an ADP-ATP pump, generating ATP from sunlight without chlorophyll. This supports the theory that chemiosmotic processes are universal in their ability to generate ATP.
CALVIN CYCLE (C3 PATHWAY)
The Calvin Cycle, earlier designated the photosynthetic "dark reactions" pathway, is now referred to as the carbon reactions pathway. In this pathway, the free energy of cleavage of ~P bonds of ATP, and reducing power of NADPH, are used to fix and reduce CO2 to form carbohydrate. Enzymes and intermediates of the Calvin Cycle are located in the chloroplast stroma, a compartment somewhat analogous to the mitochondrial matrix.
Ribulose Bisphosphate Carboxylase (RuBP Carboxylase) catalyzes CO2 fixation:
ribulose-1,5-bisphosphate + CO2 à 2 copies of 3-phosphoglycerate
Because it can alternatively catalyze an oxygenase reaction , the enzyme is also called RuBP Carboxylase/Oxygenase (RuBisCO). It is the most abundant enzyme on earth. The RuBP
|
Extraction of a proton from C3 of ribulose-1,5-bisphosphate (RuBP, below left) promotes formation of an endiolate intermediate.
|
|
|
Transition state analogs of the postulated b-keto acid intermediate bind tightly to the enzyme and inhibit its activity. Examples include 2-carboxyarabinitol-1,5-bisphosphate (CABP, at right) and carboxyarabinitol-1-phosphate (CA1P).
RuBP Carboxylase in plants is a complex (L8S8) of: |
|
Some bacteria contain only the large subunit, with the smallest functional unit being a homodimer, L2. Roles of the small subunits have not been clearly defined, although there is some evidence that interactions between large and small subunits may regulate catalysis.
|
Large subunits within RuBisCO are arranged as antiparallel dimers, with the N-terminal domain of one monomer adjacent to the C-terminal domain of the other monomer. Each active site is at an interface between monomers within an L2 dimer, explaining the minimal requirement for a dimeric structure. The substrate binding site is at the mouth of an ab-barrel domain of the large subunit. Most active site residues are polar, including some charged amino acids (e.g., Thr, Asn, Glu, Lys). |
|
|
"Active" RuBP Carboxylase includes a carbamate group, that binds an essential Mg++ at the active site. The carbamate forms by reaction of HCO3- with the e-amino group of a lysine residue of RuBP Carboxylase, in the presence of Mg++. HCO3- that reacts to form the carbamate group is distinct from CO2 that binds to RuBP Carboxylase as substrate.
The active site Mg++ bridges between oxygen atoms of the carbamate and the substrate CO2. |
|
Binding of either the normal substrate ribulose-1,5-bisphosphate or a transition state analog to RuBP Carboxylase causes a conformational change to a "closed" conformation in which access of solvent water to the active site is blocked.
RuBP Carboxylase (RuBisCO) can spontaneously deactivate by decarbamylation. In the absence of the carbamate group, RuBisCO tightly binds ribulose bisphosphate (RuBP) or another sugar phosphate at the active site as a "dead end" complex, with the closed conformation, and is inactive in catalysis. In order for the carbamate to reform, the enzyme must undergo transition to the open conformation.
RuBP Carboxylase Activase, an ATP hydrolyzing (ATPase) enzyme, causes a conformational change in RuBP Carboxylase from closed to open form. This allows release of tightly bound RuBP or other sugar phosphate from the active site, and carbamate formation. Since photosynthetic light reactions produce ATP, the ATP dependence of RuBisCO activation provides a mechanism for light-dependent activation of the enzyme.
RuBP Carboxylase Activase is a member of the AAA family of ATPases, many of which have chaperone-like roles. The activase is a large multimeric protein complex that may surround RuBP Carboxylase while inducing the conformational change to the open state.
|
The normal product of the RuBP Carboxylase reaction, 3-phosphoglycerate, is converted to glyceraldehyde-3-phosphate. |
|
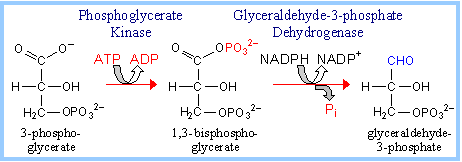
A portion of the glyceraldehyde-3-phosphate is converted back to ribulose-1,5-bisphosphate, via reactions catalyzed by Triose Phosphate Isomerase, Aldolase, Fructose Bisphosphatase, Sedoheptulose Bisphosphatase, Transketolase, Epimerase, Ribose Phosphate Isomerase, and Phosphoribulokinase. Many of these enzymes are equivalent to enzymes of the cytosolic Glycolysis, Gluconeogenesis and Pentose Phosphate Pathways, but are separate gene products resident within the chloroplast stroma. (Enzymes of the other pathways listed are located in the cytosol.) The process is similar to the Pentose Phosphate Pathway running backwards.
For three molecules of ribulose-1,5-bisphosphate (total of 15 C) that are carboxylated, cleaved, phosphorylated, reduced, and dephosphorylated, six molecules of glyceraldehyde-3-phosphate are produced (total of 18 C). Of these:
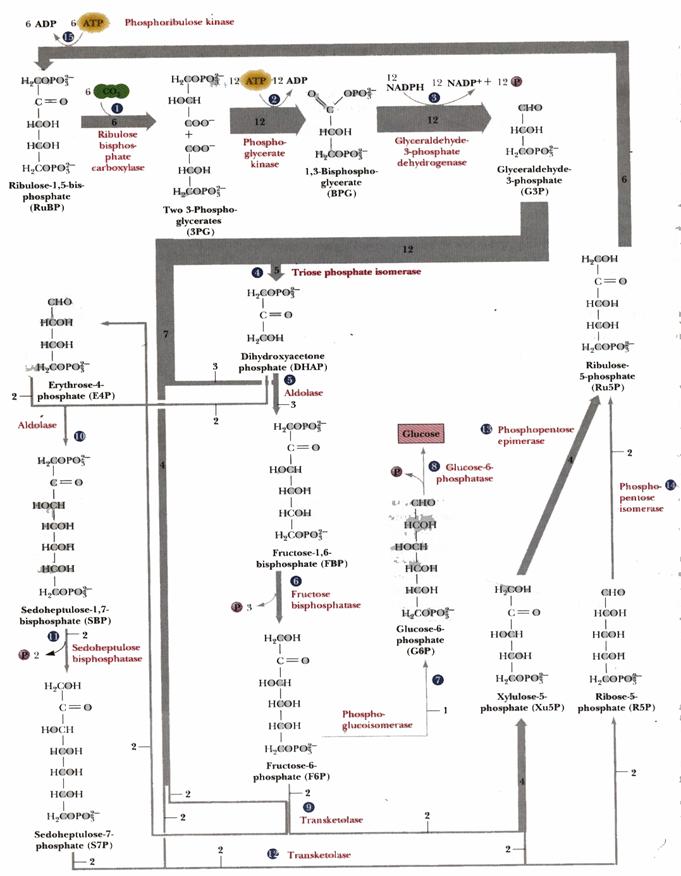
Five of the 3-C molecules (total of 15 C) are recycled back into three 5-C molecules of ribulose-1,5-bisphosphate, (substrate for RuBisCO), as summarized below.
C3 + C3
à
C6
C3 + C6
à
C5 + C4
C3 + C4
à
C7
C3 + C7
à
C5 + C5
Overall: 5C3 à 3C5
|
Enzymes in the diagram at right:
|
|
Summary of Calvin Cycle, omitting compounds that are regenerated:
3 CO2 + 9 ATP + 6 NADPH à glyceraldehyde-3-phosphate + 9 ADP + 8 Pi + 6 NADP+
|
Glyceraldehyde-3-phosphate may be converted to other carbohydrate metabolites (e.g., fructose-6-phosphate and glucose-1-phosphate), energy stores (e.g., sucrose or starch), or cell wall constituents (e.g., cellulose). Glyceraldehyde-3-P can also be utilized by plant cells as carbon source for synthesis of fatty acids and amino acids.
|
|
|
REGULATION OF CALVIN CYLE:
Regulation prevents the Calvin Cycle from being active in the dark, when it might function in a futile cycle with Glycolysis and Pentose Phosphate Pathways, wasting ATP and NADPH.
Light activates, or dark inhibits, the Calvin Cycle (previously called the "dark reaction") in several ways. |
|
Light-activated electron transfer is linked to pumping of H+ into thylakoid disks. The pH in the stroma increases to about 8. The alkaline pH activates stromal Calvin Cycle enzymes RuBP Carboxylase, Fructose-1,6-Bisphosphatase and Sedoheptulose Bisphosphatase.
The light-activated shift of H+ into thylakoid disks is countered by Mg++ release from the thylakoids to the stroma. RuBP Carboxylase (in the stroma) requires Mg++ binding to carbamate at the active site.
Some plants synthesize the transition-state inhibitor carboxyarabinitol-1-phosphate (CA1P) in the dark. RuBP Carboxylase Activase facilitates release of CA1P from RuBP Carboxylase, when it is activated under conditions of light via thioredoxin.
|
Thioredoxin is a small protein with a disulfide that is reduced in chloroplasts via light-activated electron transfer. The twisted 5-stranded b-sheet surrounded by 4 a-helices is typical of members of the thioredoxin protein family.
|
|
|
During illumination, the thioredoxin disulfide is reduced to a dithiol by ferredoxin, a constituent of the photosynthetic light reaction pathway, via an enzyme Ferredoxin-Thioredoxin Reductase. The reduced thioredoxin activates several Calvin Cycle enzymes, including Fructose-1,6-bisphosphatase, Sedoheptulose-1,7-bisphosphatase, and RuBP Carboxylase Activase, by reducing specific disulfides in these enzymes to thiols. |
The C4 pathway
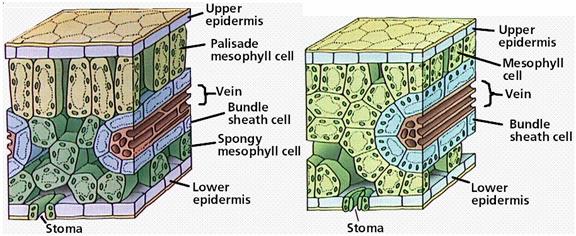
The C4 pathway is designed to efficiently fix CO2 at low concentrations and plants that use this pathway are known as C4 plants. These plants first fix CO2 into a four carbon compound (C4) called oxaloacetate. This occurs in cells called mesophyll cells. First, CO2 is fixed to a three-carbon compound called phosphoenolpyruvate to produce the four-carbon compound oxaloacetate. The enzyme catalyzing this reaction, PEP carboxylase, fixes CO2 very efficiently so the C4 plants don't need to to have their stomata open as much.
The oxaloacetate is then converted to another four-carbon compound called malate in a step requiring the reducing power of NADPH. The malate then exits the mesophyll cells and enters the chloroplasts of specialized cells called bundle sheath cells. Here the four-carbon malate is decarboxylated to produce CO2, a three-carbon compound called pyruvate, and NADPH. The CO2 combines with ribulose bisphosphate and goes through the Calvin cycle while the pyruvate re-enters the mesophyll cells, reacts with ATP, and is converted back to phosphoenolpyruvate, the starting compound of the C4 cycle. The C4 cycle is summarized in the figure.
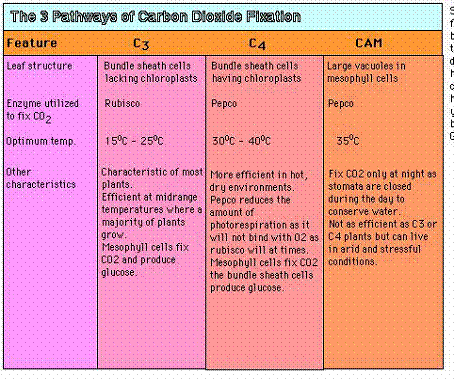
The CAM pathway:
CAM plants live in very dry condition and, unlike other plants, open their stomata to fix CO2 only at night. Like C4 plants, the use PEP carboxylase to fix CO2, forming oxaloacetate. The oxaloacetate is converted to malate which is stored in cell vacuoles. During the day when the stomata are closed, CO2 is removed from the stored malate and enters the Calvin cycle.
PHOTORESPIRATION :
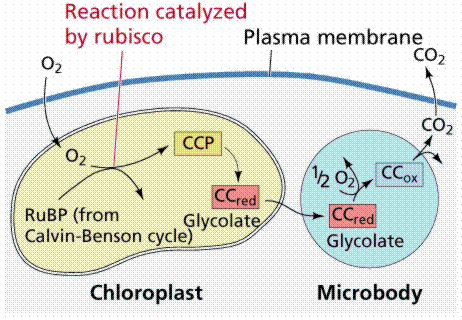
The capture of carbon dioxide by PEP is mediated by the enzyme PEP carboxylase, which has a stronger affinity for carbon dioxide than does RuBP carboxylase When carbon dioxide levels decline below the threshold for RuBP carboxylase, RuBP is catalyzed with oxygen instead of carbon dioxide. The product of that reaction forms glycolic acid, a chemical that can be broken down by photorespiration, producing neither NADH nor ATP, in effect dismantling the Calvin Cycle.
|
This photorespiration is the basis for the name RuBP Carboxylase/ Oxygenase (RuBisCO).
|
|
|
|
|
The pathway that salvages some carbon from 2-phosphoglycolate, partially converting it to 3-phosphoglycerate, uses up ATP and NADH. Photorespiration is a wasteful process, reducing efficiency of CO2 fixation by as much as 50%.
PHOSPHORYLATION
Addition of phosphorus is referred as phosphorylation. The energy released during oxidation reaction is stored in the form of high energy phosphate bonds in ATP. When energy is released, the ADP and Pi reacted and form ATP. This process of addition of Pi to from ATP is referred as phosphorylation. The high energy phosphate bonds are differing from normal phosphate bonds and the energy released from ATP is higher than glucose phosphates.
The phosphorylation or ATP formation occurs in three different ways in bacteria: