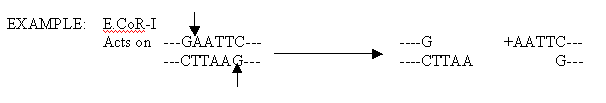
STRUCTURE OF NUCLEIC ACID
Nucleic acids are of two types namely, DNA and RNA. DNA is Deoxy Ribonucleic acid, it contains deoxy ribose sugar, phosphate and bases like adenine, guanine, thymine and cytosine whereas RNA, ribonucleic acid, contains ribose sugar, phosphate and bases like
Adenine, guanine, uracil and cytosine.
STRUCTURE OF DNA:
Structure of DNA studied under four different headings. They are
i) Primary structure
ii) Secondary structure
iii) Tertiary structure
iv) Quaternary structure
i) Primary structure:

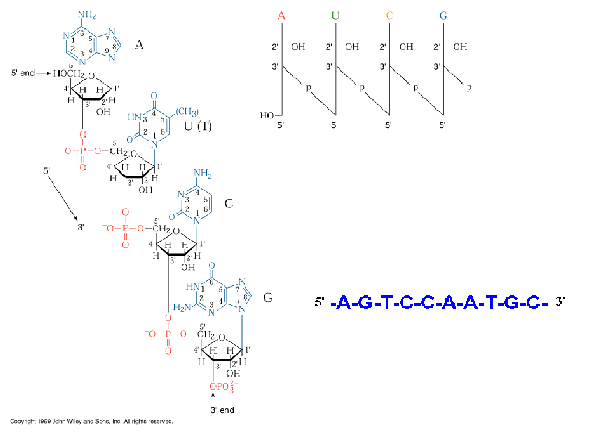
Primary structure of DNA is formed by the covalent
backbone consisting of deoxyribo nucleotides linked to each other by
phosphodiester bonds. DNAs are long
chains of nucleotide units or polydeoxyribonucleotides.
The substrates for polymerization are nucleoside triphosphates, but the
repeating unit or monomer, of a nucleic acid is a monophosphate.
During polymerization, the 3’-OH group of the terminal nucleotide
residue in the existing chain makes a nucleophilic attack upon the alpha
phosphate of the incoming nucleoside triphosphate to form 5’à3’
phosphodiesterbond. This reaction
is catalyzed by DNA polymerase.
Serial polymerization generates long polymers variously called chains or
strands, containing an invariant sugar-phosphate backbone with 5’à3’
polarity and projecting nitrogenous
base.
The most importamt clue to the DNA structure came from the discovery made by Erwin charagaff and etal in 1940s. series of experiments conducted by them lead to postulate a series of results are called as charagaffs’ rule. They are
a) DNA specimens isolated from different tissues of the same species have the same base composition.
b) Dna base composition varies from one species to another species.
c) In a given species, the base composition of DNA does not change with age, nutritional states or change in environoment
d) The number of adenine residues in all DNAs irrespective of species is equal to the number of thymine residues(A=T) and the number of guanine residues equal to the cytosine residues(G=C).
From this, the following relation derived
Purines = Pyrimideines;
A+G = T+C;
ii) SECONDARY STRUCTURE:James Watspm and Francis Crick postulated the secondary structure of DNA in 1953 in their double helical model. They studied the x-ray diffraction pattern of DNA made by Rosalind Franklin and Maurice Wilkins and arrived at the double helical structure DNA. Features of double helical structure is as follows:
i) DNA molecule present as an alpha helical structure and if it is double stranded structure. Here, the relatively hydrophobic bases are present inside the molecule which polar sugar and phosphoric acid molecules are present on the outside. This shields hydrophobic bases from water and made hydrophilic sugar and phosphoric acid molecule to interact with water.
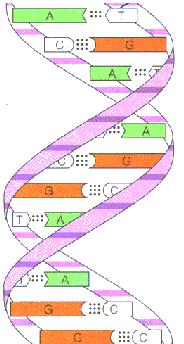
iii) Two polynucleotide chains are associated by hydrogen bonds between bases. The double helical structures is stablised by hydrophobic interactions between adjacent bases brought about by electrons in pi rings. There are two hydrogen bonds between adenine and thymine. Three hydrogen bonds present between guanosine and cyotosine.Pairing between two purines and two pyrimidine bases does not occurs. The diameter of the strands only 2nm. Two purines would be too large to be accommodate within the space. Two pyrimidines would be too small and the hydrogen bonding between two pyrimidines would be too weak.
iv) Co planarity of the bases: Bases in the same plane are straggled one above the other perpendicular to the central axis.
v) Complementarities of the bases:
The two strands are exactly complementary with each other. Thus if the sequence of bases of one strand is known, the sequence of the other strand can be easily written down.
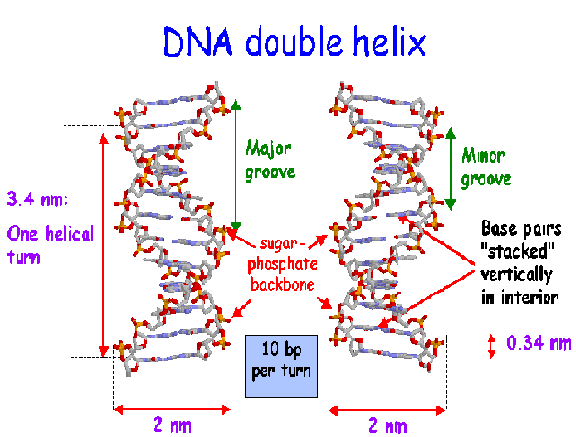
vi) Periodicity:
Ten mononucleotides are present in one turn of the helix giving periodicity of 3.4nm along the central axis of the DNA. The distance between two nucleotide is 0.34nm. the spatial relationship between the two strands create the major groove and minor groove.
vii) Anti parallel polarity: Two strands are antiparallel to each other and run in opposite direction i.e. 3’à5’ and 5’à3’
CONFORMATIONAL VARIATION IN DOUBLE HELICAL STRUCTURE:
Change in conformation provides four different kind of DNA molecules namely A –DNA, B-DNA,C-DNA and Z-DNA.The structure discovered by Watson and Crick, referred to as B-DNA, represents the sodium salt of DNA. Under highly humid conditions. DNA can assume different conformations because deoxyribose is flexible and the C-N glycosidic linkage rotates.

When DNA becomes partially dehydrated, it assumes the A form. In A-DNA the base pairs are no longer at right angles to the helical axis. Instead they like 20* away from the horizontal. In addition, the distance between adjacent base pairs is lightly reduced, with 11bp per helical turn instead of the 10.4bp found in B-form. Each turn of the double helix occurs in 2.5nm, instead of 3.4nm and the molecules’ diameter swells to approximately 2.6nm from the 2.4nm observed in B-DNA. The A form of DNA is observed when it is extracted with solvent such as ethanol. The significance of A-DNA under cellular conditions is that the structure of RNA duplexes and RNA-DNA duplexes formed during transcription resemble the A-DNA structure.
The C-DNA is found in fibers of lithium DNA at 66% relative humidity. This also a right-handed helix. This structure occurs in chromatin and some viruses. The number of residues present per helical turn is 9.3bps. the distance between tow base pairs is 0.4nm.
The Z-DNA named for its zigzag conformation radically departs from the B form. Z-DNA (diameter=1.8nm) is considerable slimmer than B-DNA. Z-DNA is twisted into a left-handed spiral with 12bps per turn. Each turn of Z-DNA occurs in 4.5nm. DNA segments with alternating purine and pyrimidine bases are most likely to adopt a Z configuration. In Z-DNA, the bases stack in a left handed staggered dimeric pattern, which gives the DNA zigzag appearance and it flattened, nongrooved surface. Regions of DNA rich in GC repeats are often regulatory, binding specific proteins that initiate or block transcription certain physiologically relevant processes such methylation and negative supercoiling stabilize the Z-form.
COMPARISON OF SOME MORPHOLOGICAL FEATURES AND SELECTED BOND TORSION ANGLES AND HELICAL PARAMETERS OF THE THREE MAJOR TYPES OF DNA HELIX:
|
FEATURES |
A |
B |
Z |
|
Morphological Characteristics: |
|
|
|
|
Helix rotation sense |
Right handed |
Right handed |
Left handed |
|
Pitch(base pairs per turn) |
~11bps |
~10bps |
~12bps |
|
Base pairs per helix repeat |
One |
One |
Two |
|
Helix axis location |
Major groove |
Through base pairs |
Minor groove |
|
Overall proportions |
Short and broad |
Longer and thinner |
Elongated and thin |
|
Major groove |
Deep, narrow |
Wide |
Flat |
|
Minor groove |
Broad, shallow |
Narrow |
Narrow, very deep |
|
Helix diameter |
2.3nm or 2.5nm |
1.9nm or 2.3nm |
1.8nm |
|
TORSIONAL PARAMETERS |
|
|
|
|
Sugar pucker |
C-2’ endo |
C-3’ endo |
Alternating |
|
Glycosidic bond conformation |
anti |
anti |
Alternating anti/syn |
|
HELICAL PARAMETERS |
|
|
|
|
Displacement |
-4.4 |
0.6 |
3.2 |
|
Twist |
33 |
36 |
-49\-10 |
|
Rise |
2.6 |
3.4 |
3.7 |
|
Inclination |
22 |
-2 |
-7 |
iii) TERTIARY STRUCTURE:
Nucleic acid tertiary structures reflect interactions which contribute to overall three dimensional shape. This includes interactions between different secondary structure elements, interactions between single strands and secondary structures and topological properties of nucleic acids. Examples for tertiary include cruciform, triple helices and super coils.
a. Cruciform:
Cruciform are cross-links structures. They are likely to form when a DNA sequence contains a palindrome or inverted repeats.(palindrome is defined as a sequence that provides the same information whether it is read forward or backward e.g. MADAM I’M ADAM). In one proposed mechanism, cruciform formation begins with a small bubble, or protocruciform and progresses as intrastrand base pairing occurs. The mechanism, which bubble initiates formation, is unknown. The function of cruciform is unclear but is believed to be associated with the binding of various proteins to DNA. DNA palindromes also play a role in the function of an important class of enzymes called the restriction enzymes.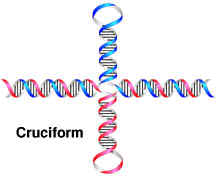
b. Triple helix:
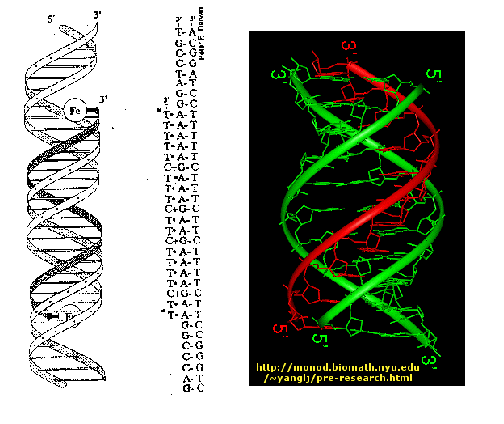
In certain circumstances e.g. low pH, a DNA sequence containing a long segments consisting of a polypurine stand hydrogen-bonded to a polypyrimidine strand can form a triple helix. The formation of the triple helix, also referred to a H-DNA, depends on the formation of nonconventional base pairing (HOOGSTEEN Base pairing), which occurs without disruption the Watson-Crick base pairs. The significance of H-DNA is unclear, although it is implicated in the regulation of some genes. Ex: GAP-43 in mammals. Triple stranded DNA also forms during recombination when a single stand invades a duplex.
Quadruple structure is one of the tertiary structure of DNA. Guanosine repeats like in telomere ex: (T4G2)2 in Oxytrichia can associate to form cyclic tetramers known as G-quartets.
c) Super coiling :
Topology is the branch of mathematics dealing with the properties of geometric structures, which are independent of size, shape and unchanged by deformation. If a double stranded DNA molecule has free ends, the two strands wind around eachother in the most energetically favorable manner and the molecule is said to be relaxed DNA. Double stranded circular DNA or linear DNA duplex whose ends are not free to rotate, form superoils if the strands are under wound(negatively supercoiled) or over wound(positively super coiled). Under wound duplex DNA has fewer than the natural number of turns whereas overwouond DNA has more.
Negative super coiling introduces a torsional stress that favors unwinding of the right handed B-DNA double helix, while positive super coiling pverwinds such a helix. Both forms of supercoiling compact the DNA so that it sediments faster upon ultra centrifugation or migrates more rapidly an electrophoretic gel in comparison to relaxed DNA.
The basic parameter characterizing supercoiled DNA is linking number(LK). Linking number is the number of times of one DNA strand wraps rouond the other in a duplex. For right-handed helix L is positive whereas for left handed helix L is negative. Linking number of relaxed DNA referred as L0
Super helical winding number is the difference between L and L0
DelL=L-L0
L=T+W
T=twisting
number is the total number of turns in a DNA molecule
W=writhing number is the number of super helical turns in DNA molecule
iv) QUARTERNARY STRUCTURE:In many structures, nucleic acids interact in trans e.g. the ribosome and spliceosome and this may be considered a quaternary level of nucleic acid structure. Nucleic acids also interact with an enormous number of proteins e.g. genome structural proteins, transcription factors, enzymes and splicing factors. Many of these proteins have a significant effect on DNA conformation. Interactions with proteins may be general or sequence specific and may involve subtle or overt changes in structure. Catenation is nothing but interlocking of DNA circles which occurs during replication. It is an example for quaternary structure of DNA.

RIBONUCLEICACID (RNA):
Three major types of RNAs are discussed below namely m-RNA, r-RNA and t-RNA.
m-RNA:
Messenger RNA serves to carry the information or message that is encoded in genes to the sites of protein synthesis in the cell, where this information is translated into a polypeptide sequence. Because m-RNA molecules are transcribed copies of the protein coding genetic units that comprise most of DNA, m-RNA is said to be “the DNA-like RNA”. All the RNAs m-RNA, t-RNA and r-RNA synthesized from DNA during transcription. t-RNA and r-RNA are not subsequently translated into proteins only m-RNA translated to proteins.
PROKARYOTIC m-RNA:
Prokaryotic m-RNA polycistronic in nature i.e. single m-RNA codes for more than one functional proteins. In between each gene spacers are present. Prokaryotic m-RNA translated immediately after transcription. prokaryotic m-RNA mostly available in complex form with ribosomes which referred as polysomes. Nascent m-RNA itself active. It does not undergo any postrancriptional modifications. Prokaryotic m-RNA doesn’t have cap, tail structures. M-RNA bound to ribosome with the help of shine-dalgernow sequence UCCUCC.
EUKARYOTIC m-RNA:
Eukaryotic m-RNA codes for only one polypeptide because of this they are referred as monocistronic m-RNA. They are synthesized in the nucleus in the form of much larger precursor molecules called heterogeneous nuclear RNA or hnRNA. HnRNA molecules contain stretches of nucleotide sequence that have no protein coding capacities. The noncoding regions of hnRNA are called intervening sequences or introns because they intervene between conoding regions which are called exons. Introns interrupt the continuity of the information specifying the aminoacid sequence of protein and must be spliced out before the message can be translated. In addition, 7-methyl guanosine residue added to the 5’-end of hnRNA, which is referred as capping. The 7-methyl guanosine residue referred as cap. 100 to 200 adenylic acid residues attached at their 3’-ends of hnRNA which is referred as tailing. The poly A residues referred as tail. After caping, splicing, tailing and methylation of eukaryotic hnRNA referred as matured m-RNA.
m-RNA will have primary
and secondary structure level alone.
RIBOSOMAL RNA (r-RNA):
r-RNA molecules are responsible for the formation of ribosomes which are the site of protein synthesis. Ribosomal RNA molecules fold into characteristic secondary structures as a consequences of intramolecular hydrogen bond interactions. The different species of rRNA are generally referred to according to these sedimentation coefficients.
PROKARYOTIC r-RNA:
There are three types of prokaryotic r-RNA. They are 5s-rRNA, 16S-rRNA
and 23S-rRNA. 5S-rRNA consists
120 nucleotides whereas 16S-rRNA and 23S-rRNA
consists 1542 nucleotides and 2904 nucleotides respectively.
5S-rRNA and 23S-rRNA
along with 31L proteins forms 50S ribosomal subunit.
16S-rRNA along with 21S proteins forms 30S ribosomal subunits.
These two subunit combines to form 70S ribosome.
EUKARYOTIC r-RNA:
There are four types of eukaryotic r-RNAs. They are 5S-rRNA, 5.8S-rRNA, 18S-rRNA and 28S-rRNA.
|
r-RNA TYPES |
NUMBER OF NUCLEOTIDES |
|
5S-rRNA |
120 NUCLEOTIDES |
|
5.8RNA |
160 NUCLEOTIDES |
|
18S-rRNA |
1874 NUCLEOTIDES |
|
28S-rRNA |
4718 NUCLEOTIDES |
18S-rRNA along with 33S proteins forms 40S ribosomal subunit. 5SrRNA, 5.8-rRNA and 28S-rRNA along with 49L proteins forms 60S ribosomal subunit. These two subunits comobined to form 80S ribosome.18S-rRNA responsible for the binding of m-RNA to 80S ribosome. 5S-rRNA responsible for the binding of t-RNA to 80S ribosome. 28S-rRNA and 5.8S-rRNA responsible for the peptidyl transferase activity during translation.Ribosomal RNAs caharacteristically contain a number of specifically modified nucleotides, including pseudouridine residues, ribothymidylic acid and methylated bases.
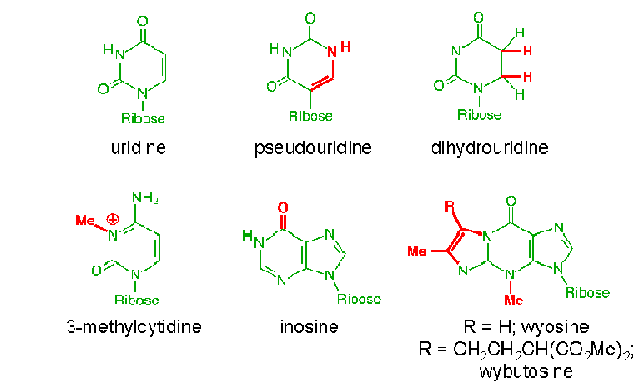
TRANSFER RNA(T-RNA):
t-RNA used to transfer amino acid residues from amino acid pool to the site of proteins synthesis. T-RNA comprising about 15% cellular RNA, the average length of t-RNA molecule is 75 nucleotides. Because each t-RNA molecule binds to a specific aminacids, cells posses at least one type of t-RNA for each of the 20 aminoacids commonly found in proteins. In practical, only 32 different t-RNA types are available. Because of this few aminoacid may have more than one t-RNA specific.
The structure of t-RNA allows it to perforom the critical functions involving the most obviously important structural components. The 3’ terminus and the anticodon loop. The anticodon loops contains a three base pair sequence that allows the t-RNA to align its attached aminoacid properly during protein synthesis. t-RNA also possess three other prominent structural features, referred to as the D-loop, the TUC loop and the variable loop. The function of these structures is unknown, but they are presumably related to the alignment of t-RNA within ribosome and the binding of a t-RNA to the enzyme that catalyzes the attachment of the appropriate aminoacid. t-RNA with aminoacid attached known as aminoacyl t-RNA synthases. The D-loop contains dihydrouridine. Similarly, the TUC loop contains the base sequence thymine, pseudouridine and cytosine. t-RNAs can be classified on the basis of the length of their variable loop. The majority of t-RNAs have variable loops with four to five nucleotides, while other have variable loops with as many as 30 nucleotides.All t-RNAs contain several ribonoucleotides that differ from the usual four for ex: 12modified bases in t-RNAphe . Only four ribonucleotides are incorporated into RNA in the transcription process. All of the rare bases found in the mature t-RNA undergoes posttranscriptional modifications.
STRUCTUTRE OF RNA:
Structure of RNA studied under different headings namely
i) Primary structure
ii) Secondary structure
iii) Tertiary structure
iv) Quaternary structure
i) PRIMARY STRUCTURE:
Ribonucleicacids(RNA) are long chains of noucleotide units or polynucleotides. The substrates ford polymerization are nucleoside triphosphates but the repeating unit or monomer, of a ribonucleicacid is a monophosphate. During polymerization, the 3’OH group of the terminal nucleotide residues in the existing chain makes a nucleophilic attack upono the alpha-phosphate of the incoming nucleoside-5’-triphosphate to form 5’à3’ phosphodiester bond. Enzymes termed RNA polymerases catalyze the reaction and pyrophosphate is produced as a byproduct. Serial polymerization generates long polymers variously called chains or strands, containing an invariant sugar-phosphate backbone with 5’à3’ polarity and projecting nitrogenous bases.
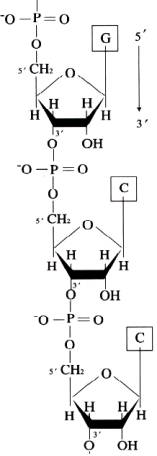
ii) SECONDARY STRUCTURE:
In RNA, secondary structure is determined by intramolecular base pairing. The major classes of intramolecular ribonucleic acid secondary structures are bulges, bulge loops, bubbles, hairpins and stem loops. A bulge is caused by a single excess residue and bulge loop by more than one. These distort stacking of neighboring bases and induce a bend , increasing the accessibility of the RNA. Internal loops or bubbles have been implicated as protein recognition site. In stem loops, the single stranded loop between the base paired helical stem may be hundreds or even thousands of nucleotides long whereas in hairpin, the short turn may contain as few as 6-8 nucleotides.
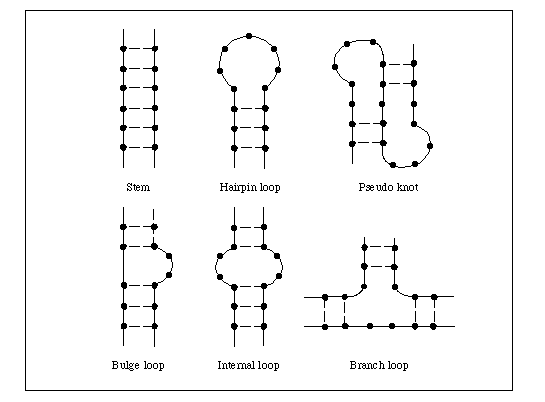
RNA secondary structures play a major role in gene expression and its regulation; base pairing between rRNA and mRNA controls the initiation of protein synthesis, base pairing between t RNA and m RNA facilitates translation, RNA hairpins and stem loops control transcriptional termination, translation efficiency and m- RNA stability and RNA- RNA base pairing also plays a major role in the splicing of introns.
iii) TERTIARY STRUCTURE:
RNA folds into complex structures involving tertiary interactions between strands, loops and duplexes. For example, in t RNA these are examples of base triples, sections of triple helix, stem junctions where two or duplex regions are joined and pseudoknots where strands interact with stem pools. Interactions between the flexible loops may result in the three dimensional folding to form pseudoknot. This tertiary structure reassembles a figure-eight knot, but the free ends do not pass through the loops, so as no knot is actually formed.
RNA folding is often controlled by molecular chaperones like protein folding. The complexity of RNA tertiary structure allows it to form biologically active molecules, and like proteins, RNA can catalyze biochemical reactions such catalytic RNAs are termed ribozymes. Some ribozymes are auto catalytic ex: the transcript of self-splicing introns, others are transacting, including ribonuclease P and the family of hammerhead ribozymes found in some plant viroids, so called because of the three helical structure of the catalytic domain.
iv) QUARTERNARY STRUCTURE:
In many structures, nucleic acid interacts in trans e.g.: the ribosome and spliceosome and this may be considered a quaternary level of nucleic acid structure. Nucleic acids also interact with a enormous number proteins e.g.: genome structural proteins, enzymes, splicing factors. Interactions with proteins may be general or sequence specific and may involve subtle or overt changes in structure.
STRUCTURE OF T-RNA:
Structure of t-RNA studied under four different headings namely
i) Primary structure
ii) Secondary structure
iii) Tertiary structure
iv) Quaternary structure
i) PRIMARY STRUCTURE:
Polymerization of ribonucleotides leads to the formation of polyribonucleotide. In polyribonucleotide, nucleotides linked by means of phosphodiester bond.
Diagram from primary structure of RNA
ii) SECONDARY STRUCTURE:
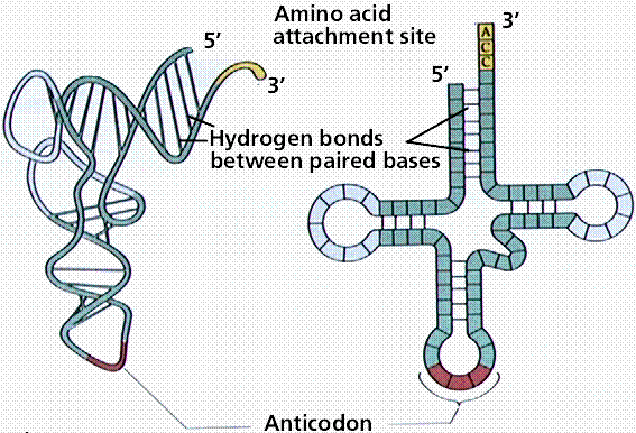
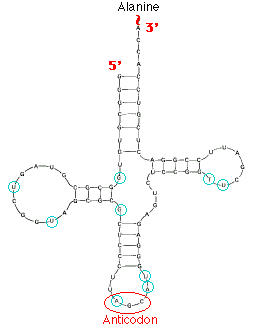
Robert Holley reported the first known base sequence of a biologically significant nucleic acid; that of yeast alanine t-RNA(t-RNAala). He proposed cloverleaf structure for t- RNA which consists of the following characters
a) 5’-terminal consists of phosphate group
b) A 7bp stem that includes the 5’ terminal nucleotide and that may contain nonwatson-crick base pairs such as G-U . This assembly is known as the acceptor or amino acid stem because the amino acid residue carried by the t- RNA is appended to its 3’terminal OH group.
c) A 4bp stem ending in a loop that frequently contains the modified base dihydrouridine. This stem and loop are therefore collectively termed as D arm,.
d) A 5bp stem ending in a loop that contains the anticodon, the triplet of bases that is comoplementary to the codon specifying the t- RNA. These features are known as the anticodon arm.
e) A 5bp stem ending in a loop that usually contains the sequence TUC. This assembly is called the TUC or T arm.
f) All t RNAs terminate in the sequence CCA with a free 3’OH group. The –CCA may be genetically specified or enzymatically appended to immature t RNA.
g) There are 15 invariant positions and 8 semi invariant positions that occur mostly in the loop regions. These regions also contain correlated invariants that is pairs of nonstem nucleotides that are base paired in all t RNAs. The purine on the 3’ side of the anticodon is invariably modified.
h) The site of greatest variability among the known t- RNAs occurs in the so called variable arm. It has from 3 to 21 nucleotides and omay have a stem consisting of upto 7bps.
i) T- RNAs also have several modified bases.
iii) TERTIARY
STRUCTURE:
In 1974, Alexander Rich separately elucidated the 2.5A resolution x-ray crystal structure of yeast t- RNAphe in collaboration with SungHoukim and in a different crystal form by Aaron Kluge. The molecule assumes an L-shaped conformation in which one leg of the L is formed by the acceptor and T stems folded into a continuous A- RNA-like double helix and the other leg is similarly composed of the D and anticodon stem. Each leg of L is 60A long and the anticodon and aminoacid acceptor sites are at opposite ends of the molecule, some 76A apart. The narrow 20 to 25A width of native t- RNA is essential to its biological function. During protein synthesis, two RNA molecules must simultaneously bind in close proximity at adjacent codons on m RNA. The bases in the nonhelical region , in the looped regions, participate in unusual hydrogen bonding interactions. These tertiary interactions may not be between bases which are complementary example A-A,G-G,A-C,G-U interaction may be seen. The ribose phosphate molecules interact with some bases in the backbone of the molecule or there may be interaction between two adjacent sugar groups. These hydrophobic interactions stabilize the molecule. The CCA terminus and adjacent helical region interact strongly with rest of the molecule.
IV) QUARTERNARY STRUCTURE:
Quarternary structure of t-RNAs formed when t-RNAs interact with proteins. These type of interaction available during protein synthesis. During translation t- RNA interact with ribosome this provides the quaternary structure for t- RNA.
PROPERTIES OF
NUCLEIC ACIDS:
A) HYPOCHROMIC EFFECT:
It has been noticed that DNA double helix absorb light in the UV region at 260nm. However, the amount of light absorbed by the native double stranded molecule is lesser than the absorption obtained by summing up the absorption of its constituents mononucleotide. This less than additive effect of the light absorption by double helix DNA is called the hypo chromic effect. It results because of the electronic interactions between the stacked bases in the native DNA double helix, which results in decreasing the amount of light each residue can absorb.
B) DENATURATION OR HELIX-COIL TRANSITION OR HYPERCHROMIC EFFECT:
Denaturation refers disruption of the native folded structure of a nucleic acid because of this, double stranded DNA molecule converted into single strands.
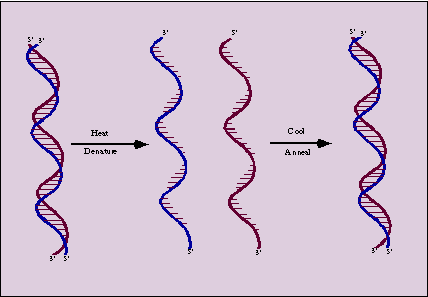
Denaturation otherwise called as helix-coil transition because during denaturation, double helix converted into single strands separates as individual random roils.Denaturation can be followed spectrophotomoetrically because the relative absorbance of the DNA solution at 260nm increases as much as 40% as the bases unstuck. This absorbance increase is referred as hyper chromic effect.
Denaturation occurs when DNA subject to
a) Extremes of temperature and pH
b) Treatment with organic solvents such as alcohol, which decrease the dielectric constant.
c)
Treatment with agents like urea, which can disrupt hydrogen bonds.
DENSITY:
During denaturation, density of DNA solution increases single stranded DNA denser than double helical DNA.
VISCOSITY:
The solution of native DNA possesses a high viscosity because of the relatively rigid double helical structure and long, rod like character of DNA. Disruption of the hydrogen bonds causes a married decreased in viscosity.
OPTICAL ROTATION:
Native DNA exhibits a strong positive rotation which is highly decreased upon denaturation. This change is analogous to the change in rotation observed when the proteins are denatured.
ABSORPTION AT 260NM:
When the native DNA is denatured, there occurs a marked increase in absorption at 260nm. This phenomenon referred as hyper chromic effect.
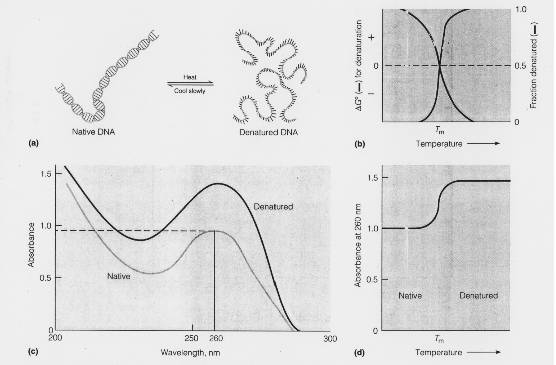
EFFECT OF HEAT ON DENATURATION:
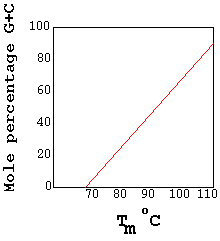
Heat can cause denaturation. If heat is used as a denaturant, the temperature at which it occurs is called melting temperature or transition temperature(Tm). Denaturation refereed as melting. The Tm depends upon the G,C content of DNA because G C pair congers extra stability to DNA double helix. If DNA has high G C content , then its Tm is high and vice versa.
EFFECT OF PH AND STRONG HYDROGEN BONDING SOLUTES:
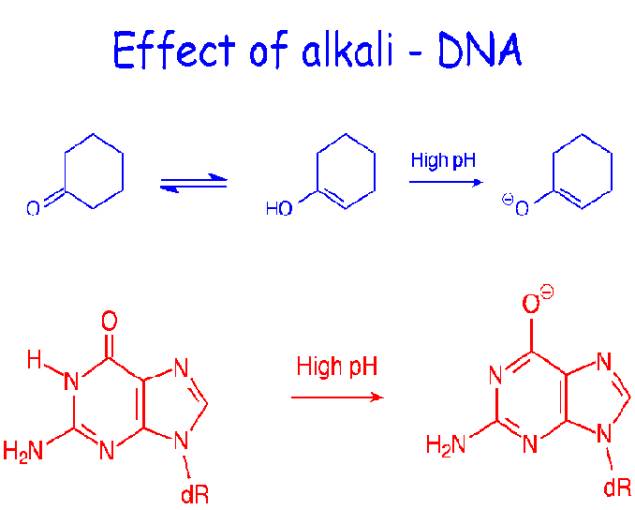
At pH a value greater than 10, extensive deprotonation of the bases occurs, destroying their hydrogen bonding potential and denaturing the DNA duplexes. Similarly, extensive protonation of the bases below pH2.3 disrupts base pairing. Alkali is preferred denaturant because, unlike acid, it does not hydrolyze the glycosidic linkages in the sugar-phosphate backbone. Small solutes that readily form hydrogen bonds are also DNA denaturants at temperatures below Tm if present in sufficiently high concentrations to compete effectively with the hydrogen bonding between the base pairs examples include formamide and urea.
C) RENATURATION
OF DNA:
Denatured dna will renature to reform the duplex structure if the denaturating conditions are removed i.e., if the solution is cooled, the pH is returned to neutrality or the denaturants are diluted out. Renaturation requires reassociation of the DNA strand into a double helix, a process termed reannealing. For this to occur, the strands must realign themselves so that their complementary bases are once again in register and the helix can be zippered up. Degree of renaturation depends on a number of factors such as:
i) Nature of the sample:
The degree of renaturation of simple sequence of DNA is high
ii) Temperature:
At low temperature(4oC) degree of renaturation is high
iii) Size of DNA fragments:
Smaller fragments show a greater degree of renaturation
iv) Ionic strength of the solution:
Too high charged DNA molecules are likely to repel each other in solutions. Hence the presence of salt necessary to mask this repulsion
v) concentration of DNA:
At higher concentration, the degree of renaturation is high
COT
VALUE:
The term cot value represents the influence of time and concentration on the renaturation of DNA. Before genome analysis by sequencing was feasible, reassociation kinetics, which is the analysis of the behavior of single, stranded nucleic acids annealing in solution was used too investigate genome properties. Although this technique is now mainly of historical interest, the principles remain useful for understanding genome architecture and nucleic acid hybridization in general. Double stranded DNA can be denatured or melted by heating and if gradually cooled will reassociate to form duplex molecules. The reassociation of single stranded DNA in solution follows second order kinetics because there are two strands and the rate at which this occurs can be expressed as follows:
![]()
where c- concentration of single stranded DNA at time T
k- the reassociation rate constant
The proportion of single stranded molecules remaining at any time, given a starting concentration of Co ,can thus be determined by integration which provide the following equation:
![]()
The above equation identifies the product of Co and t as the parameter which controls rate of reassociation. The point at which half the DNA has reassociated (t0.5) is chosen as reference. At this point C/Co = 0.5 and rearranging the above equation 2, it can be shown that Cot0.5= 1/kCot0.5. it is described as “Cot value”. Cot value is proportional to genome complexity. This is because as complexity increases, the reactive concentration of any individual sequence decreases and takes longer to find a complementary strand. The reassociation reaction thus takes longer to reach the half way point.
COT CURVES:
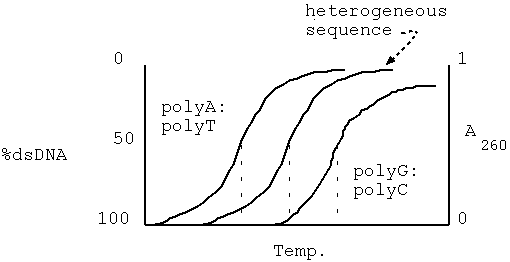
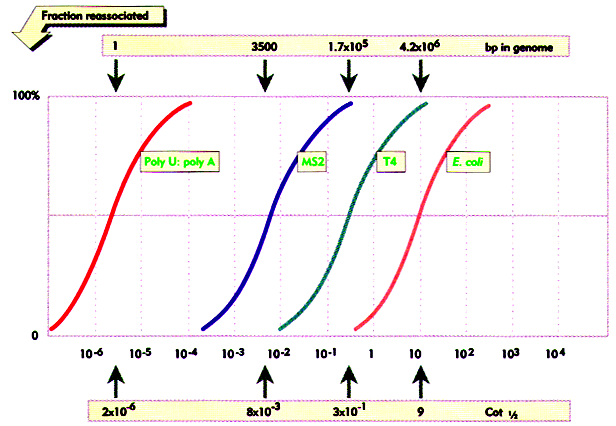
Data from genomic Cot analysis are usually plotted as log10 Cot0.5 against the fraction of reassociated DNA(1-C/Co) to give cot plot or cot curve. For the simple genomes of bacteria and viruses, reassociation occurs over two orders of magnitude of cot calues, and cot curves are linear over approximately 80% of their lengths. As the complexity of the genome increases, cot0.5 increases and curves are displaced to the right. The cot plots of E.Coli and bacteriophage lambda DNA are shown below, together with polyuridilate-polyadenylate, an artificial genome with the minimum complexity of 1. eukaryotic genomes subjected to similar analysis show reassociation over a much broader range of cot values. The eukaryotic cot plot can often be resolved into three overlapping curves, representing genome fractions with different sequence complexities. These are sometimes termed the fast, intermediate and slow components and corresponds to highly repetitive, moderately repetitive and unique sequence DNA. The slow component gives the best estimate of true genome complexity because most genes are found in unique sequence DNA. A proportion of DNA also reanneals immediately. This zero time binding DNA is also known as snapback or fold back DNA because it represents regions of dyad symmetry which can hybridize by intramolecular base pairing. The cot plot of a typical mammal is superimposed over those of the three simple genomes below
FACTORS AFFECTING DENATURATION AND RENATURATION:
|
PARAMETERS |
EFFECT ON
TM |
EFFECT ON
RENATURATION |
|
i) Base
composition |
Increase
Tm with increase in % G+C |
No effect |
|
ii) Hybrid
length |
Increase
Tm with increase in length >500bp; |
Increase
rate of renaturation with increase in length |
|
iii) Ionic
strength |
Increase
Tm with increase [Na+] |
Optimal at
1.5M[Na+] |
|
iv)
Percentage base pair mismatch |
Decrease
Tm with increase % mismatch |
Decrease
rate with increase in %
mismatch |
|
v) DNA
concentration |
No effect |
Increase
in rate with increased concentration of DNA |
|
vi)
denaturating agents |
Decrease
Tm with increasing formamide and urea |
Optimal at
50% formamide |
|
vii)
Temperature |
Not
applicable |
Optimal at
20*C below Tm |
D) NUCLEIC ACID HYBRIDIZATION:
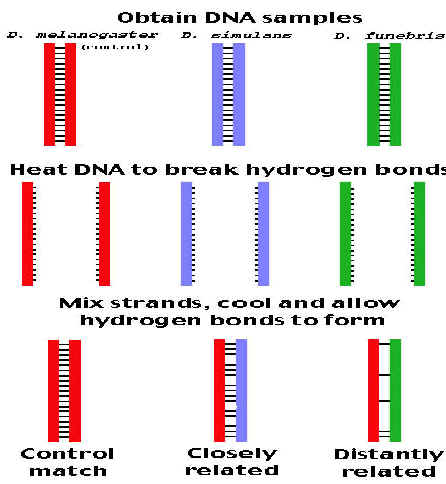
If DNA from two different species are mixed, denatured and allowed to cool slowly so that reannealing can occur, artificial hybrid duplexes (hetero duplex) may form, provided the DNA from one species is similar in nucleotide sequence to the DNA of the other. This phenomenon is referred as nucleicacid hybridization. Since DNA involved in hybridization this is otherwise known as DNA hybridization. Hybridization can occur between DNA and DNA, DNA and RNA, or RNA and RNA and may be intramolecular or intermolecular. Hybridization can occur between nucleicacids in solution or where one is in solution and the other immobilized either on a solid support or fixed insitu in a cell. Hybridization depends upon intrinsic factors and extrinsic factors. Intrinsic factors include number of hydrogen bonds, the length of duplex, its GC content and the degree of mismatch. Extrinsic factors include temperature and chemical environment.
HYBRIDIZATION IN SOLUTION:
Two DNAs from different species are heated to denature them completely, with separation of strands. When they are mixed and slowly cooled, complementary DNAs of each species will find each other and reanneal to form normal duplexes. But if the two DNAs have significant sequence homology, they will tend to form partial duplexes or hybrids with each other. The greater the sequence homology between two DNAs, the greater the number of hybrids formed. Hybrid formation can be measured by different procedures e.g. chromatography or density gradient centrifugation. Usually one of the DNAs is labeled with a radioactive isotope to simplify the measurement. This test is used to investigate sequence complexity, genome organization and gene structure.
SIMPLE FILTER HYBRIDIZATION:
Filter or membrane hybridization improves the detection of hybridized molecules by immobilizing the denatured target nucleic acid on a solid support. The transfer of nucleic acids onto such a support, which is often a nitrocellulose filter or a nylon membrane, is termed blotting. The simplest form of blotting is when the denatured sample is placed directly onto the membrane (dot plot). Alternatively, the target can be applied through a slot, which allows the area of the filter coverd, by the target to be defined refereed as slot blot. Once transferred, the nucleic acid is immobilized on the membrane. This is often achieved by baking or cross linking under UV light, although contemporary charged nylon filters bind nucleic acids spontaneously. The membrane is they incubated in a hybridization solution containing the probe and hybridization is carried out for several hours. The filter is then washed and the probe detected. This is a rapid diagnostic technique which allows the presence or absence or particular sequences to be confirmed and quantification of the target sequence.
There are four type of filter hybridization namely
|
METHOD |
COMPONENTS |
|
Southern blotting |
Separation of DNA and identification by probe |
|
Northern blotting |
Separation of RNA fragments and identification by probe |
|
Western blotting |
Separation of proteins and identification by probe |
|
Eastern blotting |
Separation of lipids and identification by probe |
REVERSE
HYBRIDIZATION:
Reverse hybridization or reverse southern or cDNA southern hybridization
involves the opposite approach of immobilizing the cloned DNA and hybridizing to
it a complex probe mixture such as labeled whole RNA or cDNA.
This technique is useful for the rapid, high expression studies where
multiple clones are tested simultaneously,e.g. to confirm that each cloned gene
is expressed in a given tissue without performing many individual hybridization.
INSITU
HYBRIDIZATION:
Insitu hybridization is the hybridizaion of a nucleic aci probe to a
target, which remain in its normal cellular location.
In this, cell or tissues are fixed, permeabilized and incubated with
probe. Advances in nonradioactive
probe technology allow the expression of several genes to be analyzed
simultaneously using different colorimetric analysis.
If fluorescent-labeled probe is used, then technique referred as Fluorescence
insitu hybridization (FISH). Application
of this technique includes cytogenetic mapping of cloned genes onto chromosomes,
the detection of virus genomes and the localization of m RNA expression. Allele
specific hybridization used to detect human disease loci in chromosomes.
E) BUOYANT DENSITY OF DNA:
G: C rich DNA has significantly higher density than A: T rich DNA. Furthermore a linear relationship exists between the buoyant densities of DNA from different sources ad their GC content. The density of DNA, as a function of its G:C content is given by the equation rho=1.660+0.098(GC) where GC= is the mole fraction of G+C in the DNA. Because of its relatively high density, DNA can be purified from cellular material by a form of density gradient centrifugation known as isopycnic centrifugation.
F) HYDROLYSIS OF
NUCLEIC ACIDS:
a) HYDROLYSI BY ACID OR BASE:
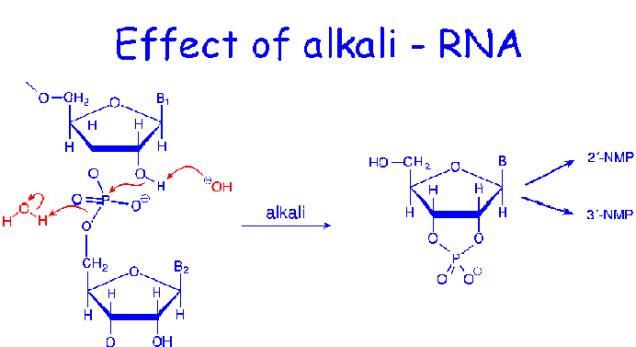
RNA is relatively resistant to the effects of dilute acid, but gentle treatment of DNA with 1mMHCl leads to hydrolysis of purine glycosidic bonds and the purine bases lost from the DNA. The purine from polynucleotide product is called apurinic acid. DNA is not susceptible to alkaline hydrolysis. On the other hand, RNA is alkali labile and is readily hydrolyzed by dilute sodium hydroxide.
Aqueous ammonia at 150*C, Ba(OH)2
NUCLEIC
ACID ![]() NUCLEOTIDES
NUCLEOTIDES
Aqueous ammonia at 175*C
NUCLEIC
ACID![]() NUCLEOSIDES+
PHOSPHORICACID
NUCLEOSIDES+
PHOSPHORICACID
NUCLEOSIDES![]() PURINES+PYRIMIDINES+SUGAR
PURINES+PYRIMIDINES+SUGAR
NUCLEICACID![]() PURINE+PYRIMIDINE+SUGAR
PURINE+PYRIMIDINE+SUGAR
12Nperchloricacid +PHOSPHORIC ACID
b) HYDROLYSIS BY ENZYME:
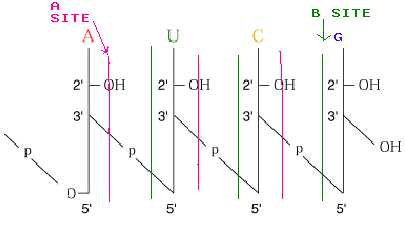
Enzyme that hydrolyze nucleic acids are called nucleases or phosphodiesterases. Because each internal phosphate in a polynucleotide backbone is involved in two phosphodiester linkages, cleavage can potentially occur on either side of the phosphorous. Convention labels the 3’-side as “a” and the 5’ side as “b”.
cleavage
on the “a” side leaves the phosphate attached to the 5’position of the
adjacent nucleotide, while b-site hydrolysis yields 3’phosphate products.
EXAMPLE: snake venom phosphodiesterase, bovine spleen phosphodiesterase
These exonucleases act on both DNA and RNA. Snake venom phosphpdiesterase act by “a” cleavage and starts at the free 3’-OH end of a polynucleotide chain, liberating nucleoside-5’-end of a nucleic acid, cleaving “b” and releasing nucleoside-3’-monophosphate.
If nuclease acts in internal location of nucleic acid they are referred as endonuclease
EXAMPLE: Pancreatic RNase – A, DNase –I, DNase – II and S1 nuclease
Pancreatic RNase specific for “b” cleavage where a pyrimidine base lies to the 3’-side of the phosphodiester bond forms oligonucleotide with pyrimidine 3’-phosphate ends.
DNase-I acts on DNA at “a” cleavage yields oligonucleotides with 3’-OH
DNase-II acts on dsDNA at “b” cleavage yields oligonucleotide
S1nuclease cleaves single stranded DNA or RNA
but not double stranded molecule at “a”
site.
Nulcease cleaving DNA referred as DNases.
Nulceases cleaving RNA referred as RNases.
RESTRICTION ENDONUCLEASES:
Restriction endonucleases are enzymes, isolated chiefly from bacteria, that have the ability to cleave double stranded DNA. The term restriction comes from the capacity of prokaryotes to defend against or restrict the possibility of takeober by foreign DNA that might gain entry into their cells. Prokaryotes degrade foreign DNA by using their unique restriction enzymes to chop it into relatively large but noninfective fragments.