GENOMIC LIBRARY CONSTRUCTION:

A genomic library is a set of recombinant clones that contain the entire DNA present in an individual organism. An E.Coli genomic library, for example, contains all the E.Coli genes, so any desired gene can be withdrawn from the library and studied. Genomic libraries can be retained for many years, and propagated so that copies can be sent from research group to research group.
Genomic libraries are prepared by purifying total cell DNA and then making a partial restriction digest, resulting in fragments that can be cloned into a suitable vector usually a cosmid or BAC or YAC. Then such recombinant vectors transformed into suitable host cells and they are cultured in suitable selective medium for recombinant vectors. Selected clones are screened for specific genes and they are labeled and maintained as library.
BAC LIBRARY:
BAC libraries are also used for cloning very large DNA fragments and have been particularly useful for sequencing large genomes. BACs are bacterial artificial chromosomes, and are based on a naturally occurring large bacterial plasmid, the F-factor. BAC vectors can accommodate DNA inserts up to 300 kb, still fairly respectable when needing to clone large genes or map and sequence complex genomes.
BAC libraries are essential elements in doing large-scale genome research such as genome sequencing, physical maps, and gene cloning. Bacterial artificial chromosomes (BAC) cloning technology was developed in early 1990s and quickly gained popularity. This technology was used in applications for genomic analysis that included large-scale physical mapping and genomic sequencing although this technology was developed much later than Yeast Artificial Chromosome (YAC) and cosmid cloning technologies. This should be attributed to the technical advantages of BAC cloning over YAC and cosmid cloning system summarized as follows:
1) Bigger insert size (up to 300 kb) than cosmid and other plasmid system.
2) Much more stable than yeast.
3) Bacterial clones and libraries grow much faster than YAC.
4) Easier in handling and gridding, DNA preparation.
5) Allows for more efficient screening by either hybridization or PCR based screening.
BAC vectors are derived from the F factor of E. coli. The F factor naturally occurs as a 100-kb molecule. Similar to shotgun cloning, BAC vectors contain the minimal sequences needed for autonomous replication, copy-number control and partitioning of the plasmid. The recombinant BAC vectors also contain an antibiotic resistant gene in addition to the blue and white colour selection system. The multiple cloning cassette in the BAC vector has sites for a dozen of frequently used restriction enzymes including but not restricted to EcoRI, HindII, NotI, BamHI, SphI
BAC Library construction can be achieved in the following steps. For library construction, initially, Genome whose library has to be prepared treated with suitable restriction enzyme which is capable of producing DNA fragments inserted into BAC. Similarly, BAC also treated with same restriction enzyme. Then in the next stage, these DNA fragments and open BAC are mixed in the presence of the enzyme DNA ligase for the formation of recombinant vector.
After suitable time interval, the recombinant vectors with DNA fragments, transformed to E.Coli with electroporation method. Transformed E.Coli culture then allowed to grow in suitable selection medium with X-gal because BAC contains beta-galactosidase gene where restriction enzyme site relies. Due to the above reason, the recombinant vector containing colonies provide white color whereas self sealed vector (nonrecombinant vector) containing colonies provide blue color due to the expression of galactose enzyme.
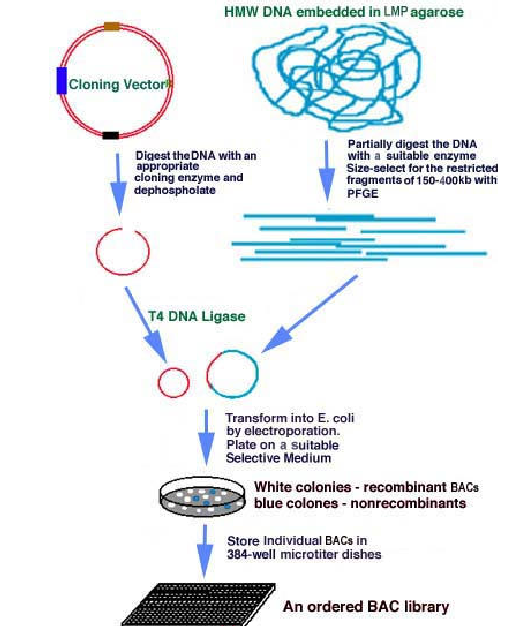
White color colonies are selected and screened for the presence of particular gene then the colonies are labelled and grown in a microtiter dishes. Since they are placed according to their order of their gene in the genome from 5’-end and with the gene tag they are known as ordered BAC library.
The major applications of BAC cloning and library construction are in large-scale physical mapping and genomic sequencing and encompass the following applications:
1) Isolation of Gene clusters for entire metabolic pathway.
The BAC libraries can be employed to scan for gene pathways in drug discovery, gene pathways in various diseases, plant genetics and microbial metabolic pathways. A variety of applications derive from construction of BAC libraries including BAC Macro arrays, gene isolation by pooled PCR, homology based hybridization using various DNA probes.
2) Prepare FISH probes
Fluorescence In Situ Hybridization (FISH) employs BAC vectors as probes to illumine and analyze tissue samples to understand chromosomal rearrangements, amplifications, deletions and aneusomies. Usually fluorescent probes for specific section(s) of a chromosome are used to label a locus of a chromosome within a cell sample where the specified genetic sequences are found.
3) Complementation for map-based gene cloning using Binary vector in plants.
BAC libraries can be used to interrogate the genetic structure of plant genomes as well as mutant genotype analysis using complementation techniques. This endows the scientist with a reliable tool to study global gene expression in plant species and to isolate individual genes.
4) Genomic walking
This allows direct genome sequencing using BAC libraries instead of sequencing directly from genomes, which is a very difficult proposition and allows study of gene structure using the entire genome.
COSMID LIBRARY:
Cosmid libraries are used for cloning genes with large introns and for sequencing larger chunks of the genome. Cosmid vectors are hybrids of plasmid and bacteriophage lambda DNA (a small ~5 kb plasmid containing the plasmid origin of replication (ori), an antibiotic resistance gene such as ampr and a suitable restriction site for cloning along with the COS sequence from phage l DNA). Because of the COS sequence, cosmid recombinants can be packaged into viral particles allowing high efficiency transformation. Since most of the phage l DNA has been discarded, it can be replaced by the DNA of interest, so long as it doesn't exceed the 50 kb limit for packaging into the viral head. The insert sizes are of the order of 35-45 kb. Since the recombinant DNA does not encode any lambda proteins, cosmids do not form viral particles (or plaques) but rather forms large circular plasmids and the colonies that arise can be selected on antibiotic plates, like other plasmid DNA transformants. Cosmid clones can be manipulated similarly, allowing ease of plasmid isolation. Since many eukaryotic genes are on the order of 30 - 40 kb, the likelihood of obtaining a DNA clone containing the entire gene sequence is increased significantly when using a cosmid library.
Cosmid Library construction can be achieved in the following steps. For library construction, initially, Genome whose library has to be prepared treated with suitable restriction enzyme which is capable of producing DNA fragments inserted into cosmid. Similarly, cosmid also treated with same restriction enzyme. Then in the next stage, these DNA fragments and open cosmid vectors are mixed in the presence of the enzyme DNA ligase for the formation of recombinant vector.
After suitable time interval, the recombinant vectors with DNA fragments, packaged into phages through invitro packaging method using cos sites of cosmids. Then recombinant vectors are transformed to E.Coli with transduction method. Transformed E.Coli culture then allowed to grow in a suitable selection medium with ampicillin antibiotic since vector contains ampicillin resistance gene. Due to the above reason, the recombinant vector containing colonies alone grew in the medium.
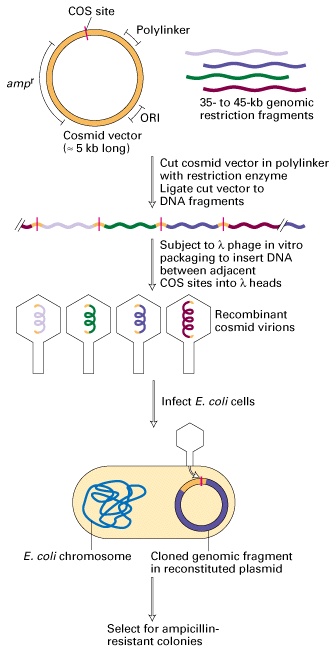
Colonies are selected and screened for the presence of particular gene then the colonies are labelled and grown in a culture plates and they are maintained for longer period.
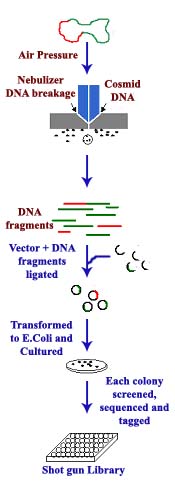
SHOTGUN LIBRARY:
Shotgun library generally prepared for sequencing genomes because sequencing methods are capable of sequencing smaller fragments. Initially genome is fragmented using nebulizer (shot gun). Fragmented DNA coupled with vectors in the presence of DNA ligase. Recombinant vectors then transformed into E.Coli and cultured in selectable medium. From the colonies grown in the culture DNA isolated from the vector and sequenced one by one. After sequencing, each colony with its genetic nature tagged and maintained as genome library.
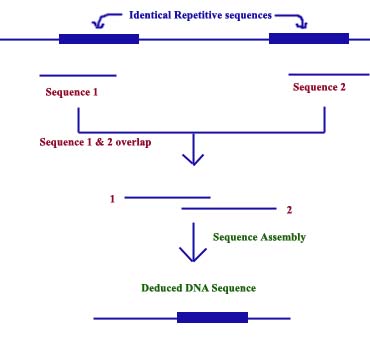
The key requirement of Shotgun approach to genome sequencing is that it must be possible to identify overlaps between all the individual sequences (DNA fragments) that are generated, and this identification process must be accurate and unambiguous so that the correct genome sequence is obtained. An error in identifying a pair of overlapping sequences could lead to the genome sequence becoming scrambled or parts missed out entirely. The probability of making mistakes increases with larger genome sizes, so the shotgun approach has been used mainly with the smaller bacterial genomes. One of the major problems in assembling genome is the presence of repetitive sequences which leads to loss of certain genes which present inbetween two repetitive sequences.
The shotgun approach for sequencing was first used successfully with the bacterium H. influenzae, which was first free-living organism whose genome was entirely sequenced, the results published in 1995. The first step was to break the 1830kb genome of the bacterium into short fragments which would provide the templates for the sequencing experiments. A restriction endonuclease could have been used but sonication was chosen because this technique is more random and hence reduces the possibility of gaps appearing in the genome sequences.
It was decided to concentrate on fragments of 1.6 – 2.0 kb because these could yield two DNA sequences, one from each end, reducing the amount of cloning and DNA preparation that was required. The sonicated DNA was therefore fractionated by agarose gel electrophoresis and fragments of the desired size purified from the gel. After cloning, 28643 sequencing experiments were carried out with 19687 of the clones. A few of these sequences (4339 in all) were rejected because they were less than 400bp in length. The remaining 24304 sequences were entered into a computer which spent 30 hours analyzing the data. The result was 140 contiguous sequences, each a different segment of the H. influenzae genome. It might have been possible to continue sequencing more of the sonicated fragments in order eventually to close the gaps between the individual segments. However, 11631485bp of sequence had already been generated which is six times the length of the genome, suggesting that a large amount of additional work would be needed before the correct fragments were, by chance, sequenced. At this stage of the project the most time effective approach was to use a more directed strategy in order to close each of the gaps individually. Several approaches were used for gap closure, the most successful of these involving hybridization analyses of a clone library prepared in a lambda vector. The library was probed in turn with a series of oligonucleotides whose sequences corresponds with the ends of each of the 140 segments. In some cases, two oligonucleotides hybridized to the same lambda clone, indicating that the two segment ends represented by those oligonucleotides lay adjacent to one another in the genome. The gap between these two ends could then be closed by sequencing the appropriate part of the lambda clone.
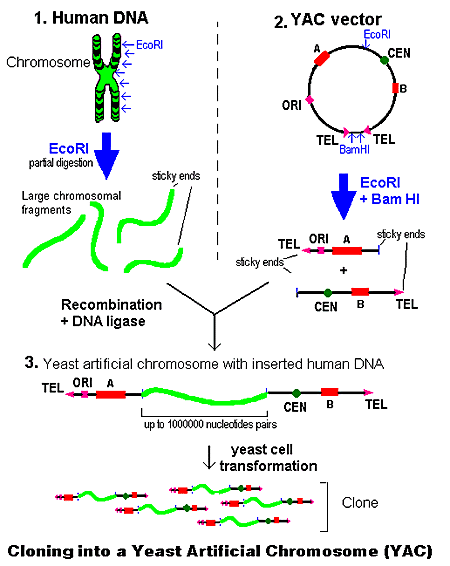
YAC LIBRARY:
YAC libraries are used for cloning very large DNA fragments (of more than 1 Mb), and are useful for cloning large genes (such as the 250 kb cystic fibrosis gene) and for creating libraries of large overlapping clones, such as for individual chromosomes isolated from organisms (chromosomal libraries). These have been used extensively for mapping genomes of complex organisms (e.g. Homo sapiens). YACs are yeast artificial chromosomes and are hybrids of bacterial plasmid DNA and yeast DNA. The components required for replication/segregation of natural yeast chromosomes have been combined with E. coli plasmid DNA. YACs are grown in the yeast Saccharomomyces cerevesiae and so contain selectable markers which are suitable for the host system. Rather than antibiotic selection, yeast selectable markers enable growth of the transformant on selective media lacking specific nutrients. (Non-transformants are unable to grow). The yeast strains that are used are auxotrophic - that is, they are unable to make a specific compound. For example, Trp1 mutants can't make tryptophan so can only grow on media supplemented with tryptophan. If the mutant strain is transformed with a YAC containing an intact Trp1 gene, then this will compensate for the inactive gene (complement) and the transfected cell is able to grow on media lacking tryptophan.
YACs are not used as extensively anymore because of inherent problems. For instance, YAC clones can contain non-contiguous segments of the genome. This means that 2 or more DNA fragments from separate parts of the genome can be integrated into an individual YAC (because they are able to support rather large inserts). A second problem is that YACs are unstable and frequently lose parts of the DNA during propagation.