IMMUNE DIVERSITY
Immune diversity is the existence of a large number of lymphocytes with different antigenic specificities in any individual. It is a fundamental property of the adaptive immune system and is the result of variability in the structures of the antigen binding sites of lymphocyte receptors for antigens i.e. antibodies from B cells and TCRs of T- cell. There are two types of immune diversity on the basis of component involved namely B cell diversity and T cell diversity.
B-CELL DIVERSITY:
The ability of B cells to produce antibodies with different specificities referred as B cell diversity. It is referred intern of Ig specificities. Polyclonal B cells can generate more than 1010 antibodies with different specificities. But each B cell clone produces Igs with single specificities.
Immunoglobins genes:
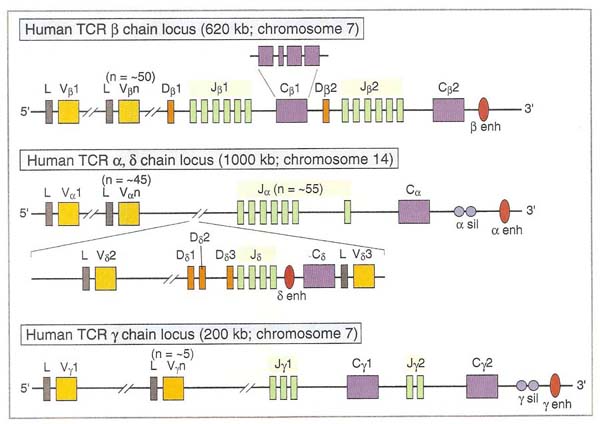
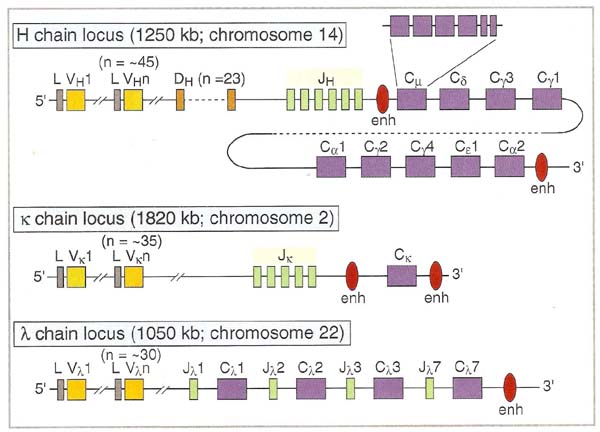
Germline Ig genes for light chain and heavy chain are shown in the following figure:

Chromosomal location of immunoglobulin genes are shown in the following table
|
S.No |
Gene |
Human Chromosome Number |
Mouse Chromosome Number |
|
1. |
k– Light chain |
2 |
6 |
|
2. |
l– Light chain |
22 |
16 |
|
3. |
H – chain |
14 |
12 |
THEORIES FOR ANTIBODY DIVERSITY:
To explain the ability of B cells to produce Igs with different specificities two theories have been proposed namely, Germline theory and somatic variation theory.
Germline theory states that the entire variable region repertoire is encoded in the germ line of the organism and is transferred from the parents to the off springs through the germ cells i.e. ovum and sperm.
Somatic variation theory states that germ line contains a limited number of variation genes which are diversified by mutation or recombinational events during development of the immune system.
Neither the germ-line nor the somatic variation theory could offer a reasonable explanation of central feature of ig structure. Germ-line proponents found it difficult to account for an evolutionary mechanism that could generate diversity in the variable part of each gene while preserving the constant region unchanged. Somatic variation proponents found it difficult to conceive of a mechanism that could diversify the variable region of a single gene in the somatic cells without allowing a single alteration in the amino acid sequence encoded by the constant region.
As an explanation for the mechanism of regulation of antibody response, Jerne postulated the network hypothesis. The variable region of an immunoglobulin molecule carrying the antigen-combining site is different antibodies. The distinct aminoacid sequences at the antigen-combining site and the adjacent parts of the variable region are termed idiotypes. The idiotype can, in turn, act as an antigenic determinant and induce anti-idiotype antibodies. These inturn can induce antibodies to them and so on, forming an idiotype network which is postulated to regulate the amount of antibodies produced and the number of antibody-forming cells in action. For this theoretical contribution to antibody formation and regulation of the immune system, Niels K. Jerne was awarded the Nobel Prize in Medicine in 1984.
In 1965, to explain antibody structure, two gene model proposed by W. Dryer and J. Bennett. They proposed that two separate genes encode a single immunoglobins heavy or light chain, one gene encoding the V region and one gene encoding the C gene region. They suggested that these two genes must somehow come together at the DNA level to form a continuous message that can be transcribed and translated to yield a single Ig heavy or light chain. Moreover, they proposed that hundreds oro thousands of V-region genes were carried in the germ line whereas only single copies of C-region class and subclass genes need exist.
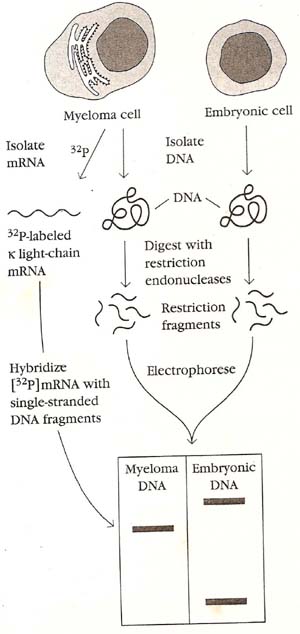
In 1976, S. Tonegawa and N. Hozumi found the first direct evidence that separate genes encode the V and C regions of immunoglobulins and that the genes are rearranged in the course of B-cell differentiation. This work changed the field of immunology. In 1987, Tonegawa was awarded the Nobel Prize for this work. Selecting DNA from embryonic cells and adult myeloma cells, Tonegawa and Hozumi used various restriction endonucleases to generate DNA fragments. The fragments were then separated by size and analyzed for their ability to hybridize with a radiolabeled mRNA probe. Two separate restriction fragments from the embryonic DNA hybridized with the mRNA, whereas only a single restriction fragment of the adult myeloma DNA hybridized with the same probe. Tonegawa and Hozumi suggested that, during differentiation of lymphocytes from the embryonic state to the fully differentiated plasma-cell stage represented in their system by the myeloma cells, the V and C genes undergo rearrangement. In the embryo, the V and C genes are separated by a large DNA segment that contains a restriction-endonuclease site; during differentiation, the V and C genes are brought closer together and the intervening DNA sequence is eliminated. This experiment is also proved with the help of southern blotting in recent years.
REARRANGEMENT:
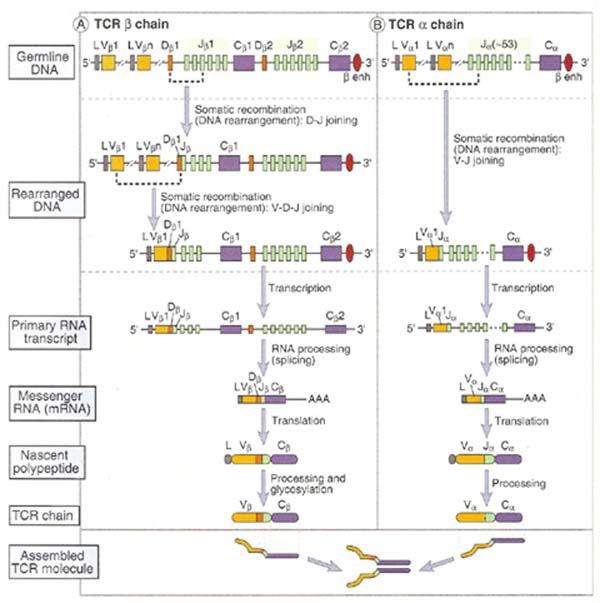
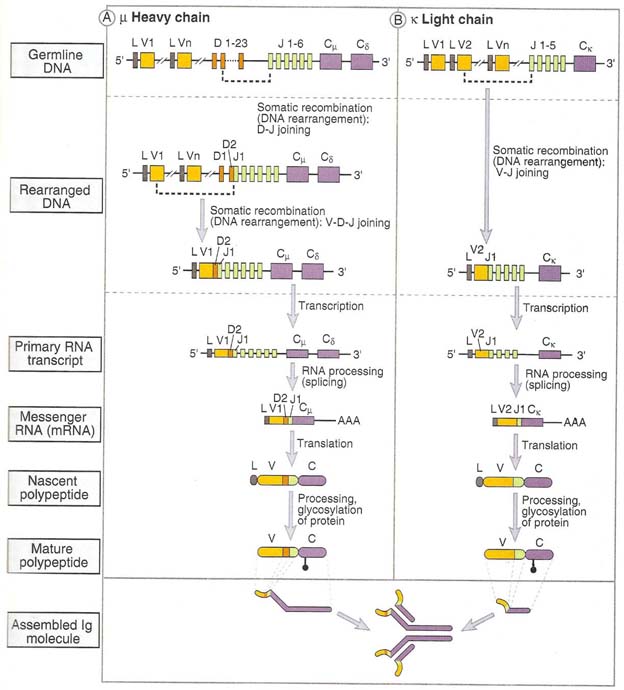
It involves the recombinational events at the DNA level to produce functional gene that encode light and heavy chains of immunoglobulins. The gene rearrangement occurs in the ordered sequence that H chain rearranges first and them L chain. This process of gene rearrangement thus leads to generation of mature, immunocompetent B cells; each such cell is antigenitically committed to a single epitope and expresses membrane bound antibody on its surface.
H – Chain rearrangement occurs in two steps in variable region. First one of the Diversity (D) regions joins to Joining (J) region and forms DJ joining. Secondly, one of the Variable (V) regions joins to DJ forming VDJ fragment. This rearranged DNA then undergoes transcription and translation to yield Heavy chain of Ig. L – Chain rearrangement occurs in one step due to the lack of diversity region. In L – chain one of the Variable (V) region selects any one of the Joining (J) region and forms VJ fragment. Then rearranged light chain undergoes transcription and translation to yield light chain of Ig.
MECHANISM FOR REARRANGEMENT:
The discovery of two closely related conserved sequences in variable-region germ-line DNA paved the way to fuller understanding of the mechanism of gene rearrangements. DNA sequencing studies revealed the presence of unique recombination signal sequences (RSSs) flanking each germ-line V, D, and J gene segment. One RSS is located 3’ to each V gene segment, 5’ to each J gene segment, and on both sides of each D gene segment. These sequences function as signals for the recombination process that rearranges the genes. Each RSS contains a conserved palindromic heptamer and a conserved AT-rich nonamer sequence separated by an intervening sequence of 12 or 23 base pairs. The intervening 12- and 23-bp sequences correspond, respectively, to one and two turns of the DNA helix; for this reason the sequences are called one-turn recombination signal sequences and two-turn signal sequences.
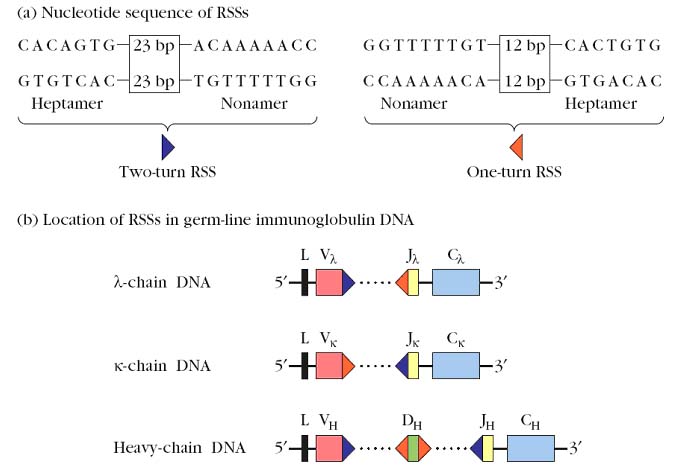
Base pair (Bp) rule states that consensus sequence with one type of spacer can be joined only to a consensus sequence with the other type of spacing and prevent one V-gene segment from recombining with other and also to J segment. Signal sequence having one turn spacer can join only with sequence having a two turn spacer called one turn / two turn joining rule.
V-(D)-J recombination, which takes place at the junctions between RSSs and coding sequences, is catalyzed by enzymes collectively called V(D)J recombinase. Identification of the enzymes that catalyze recombination of V, D, and J gene segments began in the late 1980s and is still ongoing. In 1990 David Schatz, Marjorie Oettinger, and David Baltimore first reported the identification of two recombination-activating genes, designated RAG-1 and RAG-2, whose encoded proteins act synergistically and are required to mediate V-(D)-J joining. The RAG-1 and RAG-2 proteins and the enzyme terminal deoxynucleotidyl transferase (TdT) are the only lymphoid-specific gene products that have been shown to be involved in V-(D)-J rearrangement.
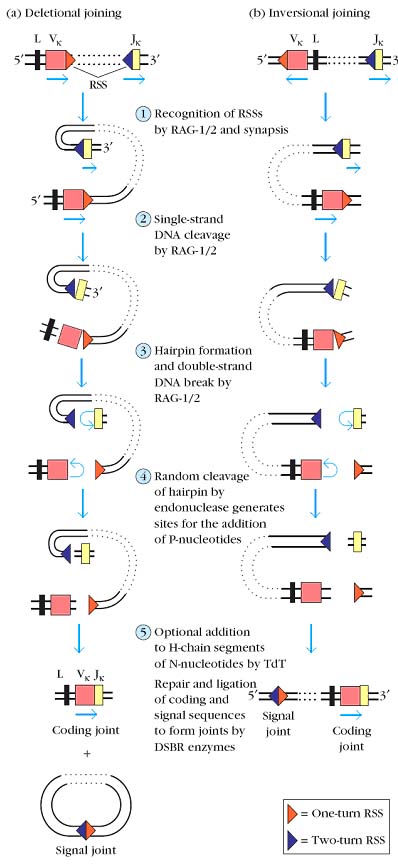
The recombination of variable-region gene segments consists of the following steps, catalyzed by a system of recombinase enzymes:
1. Recognition of recombination signal sequences (RSSs) by recombinase enzymes, followed by synapsis in which two signal sequences and the adjacent coding sequences (gene segments) are brought into proximity.
2. Cleavage of one strand of DNA by RAG-1 and RAG-2 at the junctures of the signal sequences and coding sequences.
3.A reaction catalyzed by RAG-1 and RAG-2 in which the free 3_-OH group on the cut DNA strand attacks the phosphodiester bond linking the opposite strand to the signal sequence, simultaneously producing a hairpin structure at the cut end of the coding sequence and a flush, 5_-phosphorylated, double-strand break at the signal sequence.
4. Cutting of the hairpin to generate sites for the addition of P-region nucleotides, followed by the trimming of a few nucleotides from the coding sequence by a single strand endonuclease.
5. Addition of up to 15 nucleotides, called N-region nucleotides, at the cut ends of the V, D, and J coding sequences of the heavy chain by the enzyme terminal deoxynucleotidyl transferase.
6. Repair and ligation to join the coding sequences and to join the signal sequences, catalyzed by normal double strand break repair (DSBR) enzymes.
Recombination results in the formation of a coding joint, falling between the coding sequences, and a signal joint, between the RSSs. The transcriptional orientation of the gene segments to be joined determines the fate of the signal joint and intervening DNA. When the two gene segments are in the same transcriptional orientation joining results in deletion of the signal joint, and intervening DNA, as a circular excision product.
Less frequently, the two gene segments have opposite orientations. In this case joining occurs by inversion of the DNA, resulting in the retention of both the coding joint and the signal joint and intervening DNA on the chromosome. In the human _ locus, about half of the Vk gene segments are inverted with respect to Jk and their joining is thus by inversion.
When variety of cell types were tested only preB-cells and preT-cells were able to rearrange the V and J gene segments. Mature B and T cells are unable to rearrange the VJ segments due to the lack of RAG1 and RAG2.
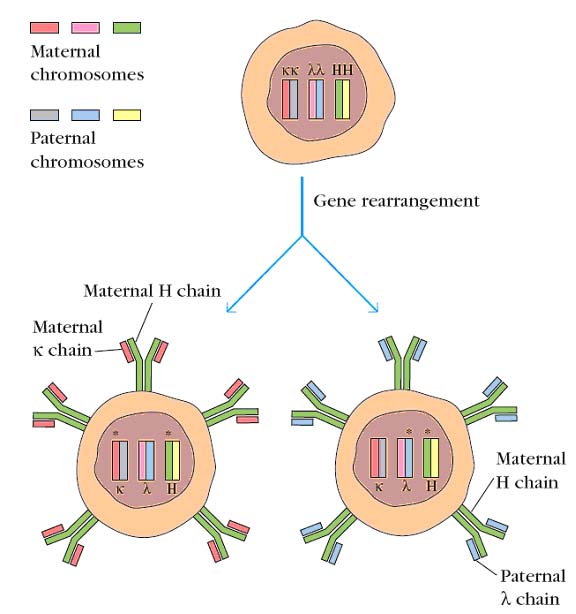
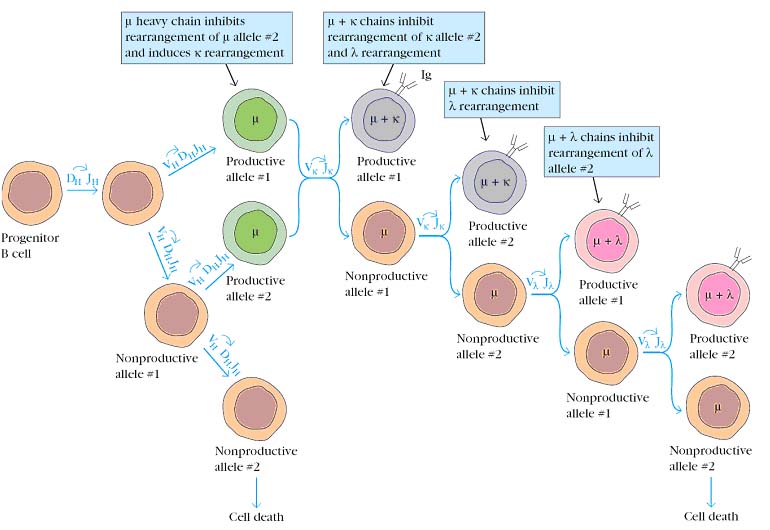
ALLELIC EXCLUSION:
B cells, like all somatic cells, are diploid and contain both maternal and paternal chromosomes. Even though a B cell is diploid, it expresses the rearranged heavy-chain genes from only one chromosome and the rearranged light-chain genes from only one chromosome. The process by which this is accomplished, called allelic exclusion, ensures that functional B cells never contain more than one VHDHJH and one VLJL unit. This is, of course, essential for the antigenic specificity of the B cell, because the expression of both alleles would render the B cell multispecific. The phenomenon of allelic exclusion suggests that once a productive VH-DH-JH rearrangement and a productive VL-JL rearrangement have occurred, the recombination machinery is turned off, so that the heavy- and light-chain genes on the homologous chromosomes are not expressed.
PRODUCTIVE AND NONPRODUCTIVE REARRANGEMENTS:
G.D.Yancopoulos and F.W.Alt have proposed a model to account for allelic exclusion. They suggest that once a productive rearrangement is attained, its encoded protein is expressed and the presence of this protein acts as a signal to prevent further gene rearrangement. According to their model, the presence of m heavy chains signals the maturing B cell to turn off rearrangement of the other heavy-chain allele and to turn on rearrangement of the k-light-chain genes. If a productive k rearrangement occurs, k light chains are produced and then pair with m heavy chains to form a complete antibody molecule. The presence of this antibody then turns off further light-chain rearrangement. If k rearrangement is nonproductive for both k alleles, rearrangement of the l-chain genes begins. If neither l allele rearranges productively, the B cell presumably ceases to mature and soon dies by apoptosis.
B CELL DIVERSITY OR ANTIBODY DIVERSITY:
There are about seven different ways in which diversity is attained in B cells. They are
1. Multiple germ gene segments:
An inventory of functional V, D, and J gene segments in the germ-line DNA of one human reveals 51 VH, 25 D, 6 JH, 40 V_, 5 J_, 31 V_, and 4 J_ gene segments. Multiple germ-line V, D, and J gene segments clearly do contribute to the diversity of the antigen-binding sites in antibodies. Due to the presence of more gene segment more possibilities available.
2. Combinatorial VDJ Joining:
The contribution of multiple germ-line gene segments to antibody diversity is magnified by the random rearrangement of these segments in somatic cells. It is possible to calculate how much diversity can be achieved by gene rearrangments. In humans, the ability of any of the 51 VH gene segments to combine with any of the 27 DH segments and any of the 6 JH segments allows a considerable amount of heavy-chain gene diversity to be generated 51 X 27 X 6 = 8262 possible combinations. Similarly, 40 Vk gene segments randomly combining with 5 Jk segments has the potential of generating 200 possible combinations at the k locus, while 30 Vl and 4 Jl gene segments allow up to 120 possible combinations at the human l locus. Thus combinatorial VDJ Joining results in B cell diversity.
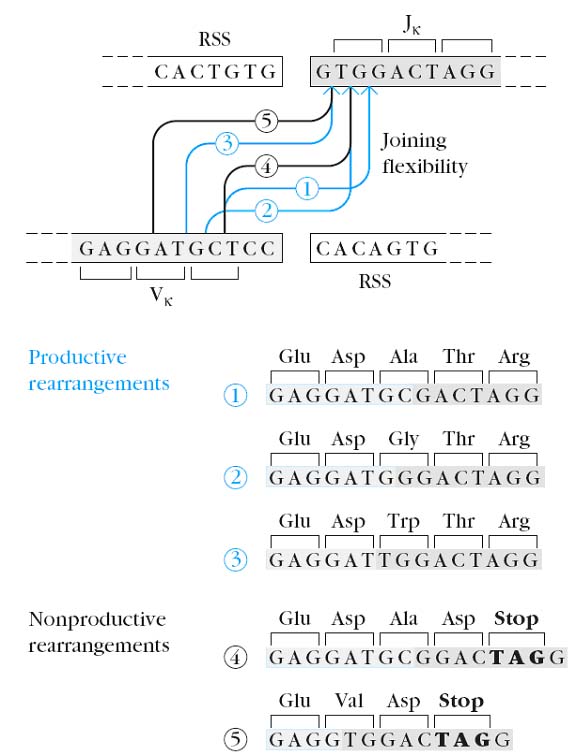
3. Junctional Flexibility:
Junctional flexibility plays an important role in diversity during recombination of signal sequences. The signal sequences are always joined precisely but the joining of coding sequences is imprecise or flexible joining which leads to diversity. The amino acid sequence variation generated by Junctional flexibility in the coding joints has been shown to fall within the third hypervariable region (CDR3) in immunoglobulin heavy-chain and light-chain DNA. Since CDR3 often makes a major contribution to antigen binding by the antibody molecule, amino acid changes generated by junctional flexibility are important in the generation of antibody diversity.
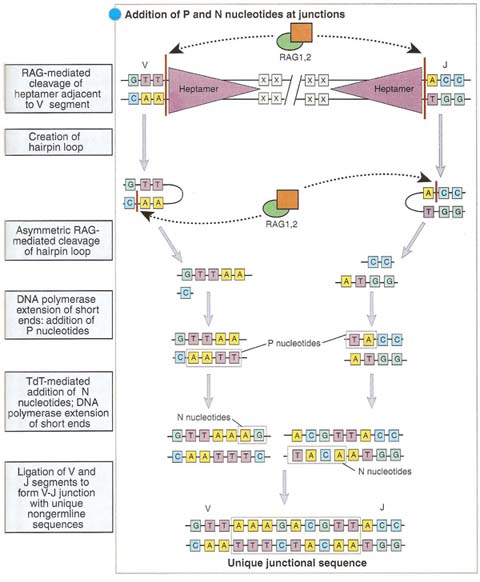
4. P-region Nucleotide Addition:
As described earlier, after the initial single-strand DNA cleavage at the junction of a variable-region gene segment and attached signal sequence, the nucleotides at the end of the coding sequence turn back to form a hairpin structure. This hairpin is later cleaved by an endonuclease. This second cleavage sometimes occurs at a position that leaves a short single strand at the end of the coding sequence. The subsequent addition of complementary nucleotides to this strand (P-addition) by repair enzymes generates a palindromic sequence in the coding joint, and so these nucleotides are called P-nucleotides. Variation in the position at which the hairpin is cut thus leads to variation in the sequence of the coding joint.
5. N-region Nucleotide Addition:
Nucleotides added during the D-J and V to D-J joining process by a terminal deoxynucleotidyl transferase (TdT) catalyzed reaction. Up to 15 N-nucleotides can be added to both the DH-JH and VH-DHJH joints. Thus, a complete heavy-chain variable region is encoded by a VHNDHNJH unit. The additional heavy chain diversity generated by N-region nucleotide addition is quite large because N regions appear to consist of wholly random sequences. Since this diversity occurs at V-D-J coding joints, it is localized in CDR3 of the heavy-chain genes. Hence, N-region nucleotide addition results in diversity.
6. Somatic Hypermutation:
Additional antibody diversity is generated in rearranged variable-region gene units by a process called somatic hypermutation. As a result of somatic hypermutation, individual nucleotides in VJ or VDJ units are replaced with alternatives, thus potentially altering the specificity of the encoded immunoglobulins. Normally, somatic hypermutation occurs only within germinal centers, structures that form in secondary lymphoid organs within a week or so of immunization with an antigen that activates a T-cell-dependent B-cell response. Somatic hypermutation is targeted to rearranged V regions located within a DNA sequence containing about 1500 nucleotides, which includes the whole of the VJ or VDJ segment. Somatic hypermutation occurs at a frequency approaching 10--3 per base pair per generation. Most of the mutations are nucleotide substitutions rather than deletions or insertions. Somatic hypermutation introduces these substitutions in a largely, but not completely, random fashion. The number of mutations progressively increased following primary, secondary, and tertiary immunizations.
7. Combinatorial diversity of H and L chains:
The specificity of an antigen binding site of an antibody is determined by the variable region of both light and heavy chain. Hence combinatorial association of heavy and light chains can generate diversity. In humans, there is the potential to generate 8262 heavy chain genes and 320 light-chain genes as a result of variable region gene rearrangements. Assuming that any one of the possible heavy-chain and light-chain genes can occur randomly in the same cell, the potential number of heavy- and light-chain combinations is 2,644,240. This number is probably higher than the amount of combinatorial diversity actually generated in an individual, because it is not likely that all
VH and VL will pair with each other. Furthermore, the recombination process is not completely random; not all VH, D, or VL gene segments are used at the same frequency. Some are used often, others only occasionally, and still others almost never.
Genetic basis of antibody diversity:
The genetic basis of antibody diversity has been clarified recently. An individual has the capacity to produce an estimated 108 different antibody molecules. To have each such antibody molecule to be coded for by a separate gene would require millions of genes to be set apart for antibody production alone. This would be obviously impossible. The phenomenons of split genes explain this. The genetic information for the synthesis of an immunoglobins molecule is not present in a continuous array of codons. Instead, this information occurs in several discontinuous stretches of DNA, each coding for separate regions of the antibody molecule. As the constant regions are identical for immunoglobulins of any one type, there need be only one gene or a few genes foreach constant region, as against a very large number of genes for the variable regions. For example, the kappa L chain genes are composed of three separate segments V, J and C. There are about a hundred different types of V (variable) domain sequences and only one C (constant) segment, with some five J (joining) segments in between. By combining different V and J sequences with the C domain. It is possible to provide for antibodies with at least 500 different specificities. By palindromic arrangement i.e. sequences that can be attached by either end, it is possible to generate many times more different specificities. The lambda chain has additional C sequences. The H chain gene has also a D (diversity) segment. By the shuffling of these different segments of the C and H chains, it is possible to have antibodies with far more than 108 types of specificity. The split gene shuffling takes place during cell development and a mature B cell DNA will have only one combination of the different segments of the immunoglobulin gene and therefore can produce only one type of antibody.
The discovery of split genes for immunoglobulins demolished the longstanding “one gene – one protein dogma in genetics and has important implications in biology, beyond immunology. For this discovery, Susumu Tonegawa was awarded Nobel prize in Medicine in 1987.
CLUSTER OF DIFFERENTIATION (CD):
A number of surface antigens or markers have been identified on lymphocytes and other leucocytes by means of monoclonal antibodies. These markers reflect the stage of differentiation and functional properties of the cells. As they were given different designations by the investigators who prepared the antibodies, the same marker came to be known by different names e.g., T4, T3, Thy1 etc. Order was introduced at the “International workshops for Leucocyte differentiation Antigens” by comparing the specificities of the different antisera. When a cluster of monoclonal antibodies were found to react with a particular antigen, it was defined as a separate marker and given a CD (Cluster of Differentiation) number. Over 150 CD markers have been identified so far. In spite of the CD nomenclature, some popular old names continue to be in use. For example, T4 and T8 is sill in use for CD4 and CD8 T cells respectively. CD notations generally used to identify and differentiate immune cells.
T-CELL DIVERSITY:
Like B cell diversity, T cell diversity depends upon the arrangement of genes for TCR. Like Heavy and light chains in B cell, TCR contains b and a chains mainly and d and g chains rarely. DNA rearrangments are same as in immunoglobulins. T cell diversity was achieved by six ways except of somatic hypermutation in B cell diversity. A mechanism for T cell diversity is same as in B cell diversity.