IMMUNOGLOBULINS
Introduction
The basic immunoglobulin structure as exemplified by IgG is a bilaterally symmetrical tetramer of two pairs of identical polypeptide chains held together by interchain disulfide bonds. Fragmentation of the molecule with enzymes made it possible to determine the structure of the molecule and gave clear indications of the relationship between its structure and function. The nomenclature (system of names) of these fragments makes it possible to talk specifically about where various functions of immunoglobulin are located. IgM and IgA are multimers of the basic structure and are held together by J chain. IgA in secretions also has secretory component which stabilizes and protects it from digestion. Comparison of amino acid sequences reveals internal similarities as well; i.e., there appears to be a repeating unit of about 110 amino acids. The amino-terminal unit in both heavy and light chains is relatively variable, but the remaining portion of both chains is more or less invariant within subclasses. This observation led to the discovery that several genes (three for light chains and four for heavy chains) are brought together to form the "gene" which subsequently transcribed into RNA and which, in turn, is translated into the light or heavy chains respectively.
Antibodies protect the host against potential parasites by: 1) directly inhibiting binding sites of viruses or various enzymes and toxins produced by bacteria, 2) agglutination, 3) opsonization which facilitates removal by phagocytes, 4) lysis of susceptible organisms via complement fixation, and 5) inducing inflammation.
The function of antibodies is to bind to antigens; thereby, directly inactivating them or activating other nonspecific systems to destroy the potential pathogens. IgG is the most important antibody in terms of its concentration, longevity, opsonic activity, ubiquity and ability to activate complement. IgM is the best at activating complement and aggregation of targets, but it is confined to the intravascular spaces except during inflammatory responses. However, it is the first immunoglobulin produced during an immune response and, along with IgD, serves as the antigen receptor for "virgin" B cells. IgA is the antibody of extracorporeal fluids and acts mainly by aggregating the potential pathogen. IgE is the antibody of allergic responses but is also an important agent of host defense against parasites.
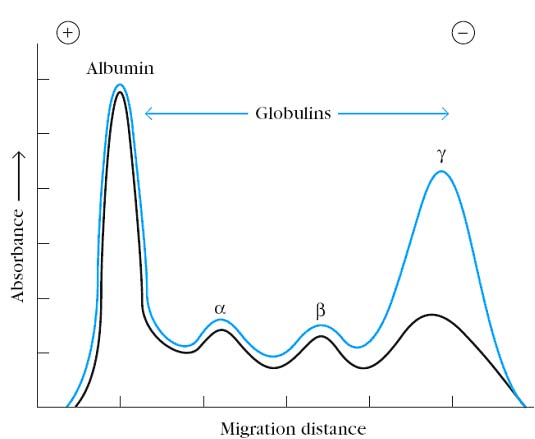
Blood can be separated in a centrifuge into a fluid and a cellular fraction. The fluid fraction is the plasma and the cellular fraction contains red blood cells, leukocytes, and platelets. Plasma contains all of the soluble small molecules and macromolecules of blood, including fibrin and other proteins required for the formation of blood clots. If the blood or plasma is allowed to clot, the fluid phase that remains is called serum. It has been known since the turn of the century that antibodies reside in the serum. The first evidence that antibodies were contained in particular serum protein fractions came from a classic experiment by A. Tiselius and E. A.Kabat, in 1939. They immunized rabbits with the protein ovalbumin (the albumin of egg whites) and then divided the immunized rabbits’ serum into two aliquots. Electrophoresis of one serum aliquot revealed four peaks corresponding to albumin and the alpha (a), beta (b), and gamma (g) globulins. The other serum aliquot was reacted with ovalbumin, and the precipitate that formed was removed; the remaining serum proteins, which did not react with the antigen, were then electrophoresed. A comparison of the electrophoretic profiles of these two serum aliquots revealed that there was a significant drop in the g-globulin peak in the aliquot that had been reacted with antigen. Thus, the g-globulin fraction was identified as containing serum antibodies, which were called immunoglobulins, to distinguish them from any other proteins that might be contained in the g-globulin fraction.

Nomenclature
Heavy chains are named by using the Greek letter that corresponds to the class name. Thus, the H chain of IgG is the gamma chain. The immunoglobulin molecule can be split into functional parts by various enzymes. This was important in establishing both the structure of the molecule and in assigning various functions to the different regions. In the 1950s and 1960s experiments by Rodney Porter and by Gerald Edelman elucidated the basic structure of immunoglobulin molecule which leads to get Nobel Prize in 1972. Porter cleaved the antibody molecule with enzymes to obtain fragments whereas Edelman dissociated the molecule by reducing the interchain disulfide bonds. The result attained by these two approaches complemented each other and allowed the basic structure of the antibody molecule to be elucidated.
Using ultracentrifugation, both Porter and Edelman first separated the g-globin fraction of serum into a high molecular weight fraction with a sedimentation constant of 19S and a low molecular weight fraction with a sedimentation constant of 7S. They used the 7S fraction containing a 1, 50,000 MW g-globin designated as Ig G for their studies.
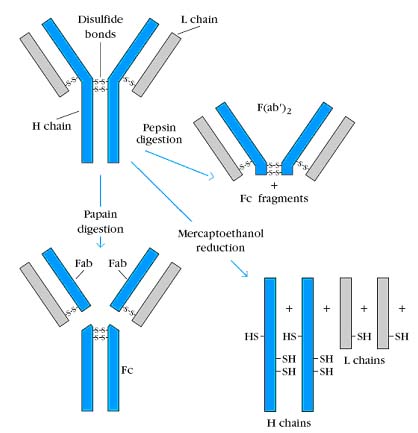
Porter subjected Ig G to brief digestion with the enzyme papain and separated the fragments. Although papain has general, nonspecific proteolytic activity and will eventually digest the entire Ig G molecule, brief treatment cleaves only the most susceptible bonds. Papain digestion of Ig G produced two identical fragments called Fab fragments, because they retained their antigen binding activity, and one fragment called the Fc fragment because it was found to crystallize during cold storage.
Similar experimental approach, but with the enzyme pepsin, was taken by Alfred Nisonoff. Brief pepsin digestion generated a single 1, 00,000 MW fragment composed of two Fab-like fragments and designated F(ab’)2. Like the Fab fragments, the F(ab’)2 fragment was also able to visibly precipitate antigens. However, after pepsin digestion, the Fc fragment was not recovered because it had been digested into multiple fragments. Porter subjected Ig G to mercaptoethanol reduction and alkylation, a chemical treatment that irreversibly cleaves disulfide bonds. The sample was then chromatographed on a column that separates molecules on the basis of size. This experiment revealed that the 1,50,000 MW Ig G molecule was composed of two 50,000 MW polypeptide chains, designated as heavy (H) chains and two 25,000 MW chains, designated as light (L) chains.
The remaining puzzle was to determine how the enzyme digestion products Fab, F(ab’)2 and Fc were related to the heavy chain and light chain reduction products. Porter answered this question by using antisera from goats that had been immunized with the Fab fragments and Fc fragments of rabbit Ig G. He found that antibody to the Fab fragment could react with both the H and L chains whereas antibody to the Fc fragment reacted only with the H chain. These observations led to the conclusion that Fab consists of portions of a heavy and a light chain and that Fc contains only heavy chain components. Based on these results, Porter and Edelman proposed the prototype structure for Ig G. According to this model, the Ig G molecule consists of two identical H chains and two identical L chains, which are linked by disulfide bridges. Papain splits the heavy chains just on the amino terminal side of the disulfide bonds holding the heavy chains together thereby producing three equal sized fragments. Two of the fragments are identical and retain the ability to bind to antigens. They are called Fab (Fragment antigen binding). The other fragment will crystallize if left in the cold overnight; thus it is called Fragment crystallizable or Fc. Another enzyme, pepsin, cleaves the molecule on the other side of the disulfide bonds (and several other places within the Fc portion). This leaves one large antigen binding fragment similar to two Fabs but with a few extra amino acids around the disulfide bond region. This fragment is called F(ab')2. The heavy chain portion of the Fab is called Fd. The area around the inter-H chain disulfide bonds is somewhat flexible, so it is called the hinge region.
Immunoglobulin Sequencing:
Ig sequencing is possible only because of using products of multiple myeloma condition, i.e., a caner of antibody producing plasma cells. The antibody produced by these lymphoma cells utilized for Ig sequencing. In these patients, light chains of Ig were discovered in the urine and were named Bence-Jones Proteins for their discoverer. Multiple myeloma also occurs in other animals. In mice it can arise spontaneously, as it does in humans or can be induced by injecting mineral oil into the peritoneal cavity. The clones of malignant plasma cells that develop are called plasmacytomas and are designated MOPCs, denoting the mineral oil induction of plasmacytoma cells. These MOPC lines utilized for the sequencing of Immunoglobins.
Light chain sequencing showed that the amino terminal half of the chain, consisting of 100 – 110 amino acids was found to vary among different Bence Jones proteins. This region was called the variable (V) region. The carboxy terminal half of the molecule, called the constant (C) region, had two basic amino acid sequences which were designated as kappa (k) and Lambda (l). In humans 60% of the light chains are kappa and 40% are lambda whereas in mice 95% of the light chains are kappa and only 5% are lambda. A single antibody molecule contains either kappa chains or lambda chains but never both. In mice there are three subtypes and in humans four subtypes of lambda present.
The amino terminal of heavy chain consisting of 100 – 110 amino acids, showed great sequence variation from one myeloma heavy chain to the next and was therefore called the variable (V) region. The remaining part of the protein revealed five basic amino acid sequence patterns corresponding to five different heavy chain constant (C) regions. Each of the different heavy chain constant region sequence is called an isotype. The length of the constant region is approximately 330 amino acids for gamma, delta and alpha and 440 amino acids for mu and epsilon. In humans there are two subclasses of alpha heavy chains and four subclasses of gamma chains.
FINE STRUCTURE:
The structure of the immunoglobulin molecule is determined by the primary, secondary, tertiary, and quaternary organization of the protein. The primary structure, the amino acid sequence, accounts for the variable and constant regions of the heavy and light chains. The secondary structure is formed by folding of the extended polypeptide chain back and forth upon itself into an antiparallel b pleated sheet. The chains are then folded into a tertiary structure of compact globular domains, which are connected to neighboring domains by continuations of the polypeptide chain that lie outside the b pleated sheets. X-ray crystallographic analysis revealed that immunoglobulin domains are folded into a characteristic compact structure called the immunoglobulin fold. This structure consists of a “sandwich” of two b pleated sheets, each containing antiparallel b strands of amino acids, which are connected by loops of various lengths. The b strands within a sheet are stabilized by hydrogen bonds that connect the –NH groups in one strand with carbonyl groups of an adjacent strand. The b strands are characterized by alternating hydrophobic and hydrophilic amino acids whose side chains are arranged perpendicular to the plane of the sheet; the hydrophobic amino acids are oriented toward the interior of the sandwich, and the hydrophilic amino acids face outward. The two b sheets within an immunoglobulin fold are stabilized by the hydrophobic interactions between them and by the conserved disulfide bond. Finally, the globular domains of adjacent heavy and light polypeptide chains interact in the quaternary structure, forming functional domains that enable the molecule to specifically bind antigen and, at the same time, perform a number of biological effector functions.
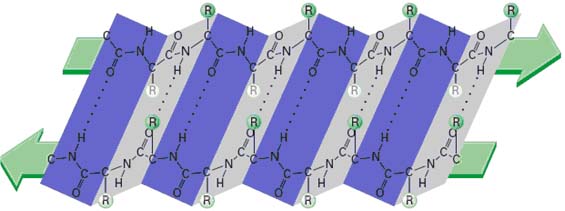
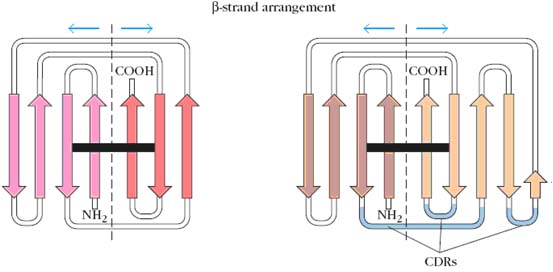
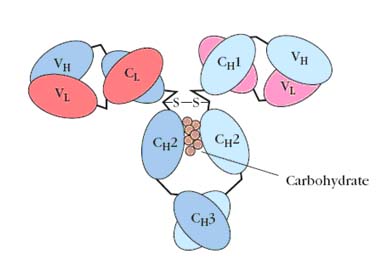
COMPLEMENTARITY DETERMINING REGION (CDR):
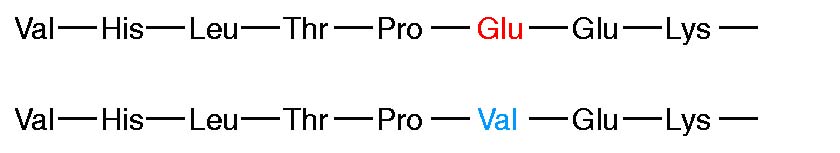
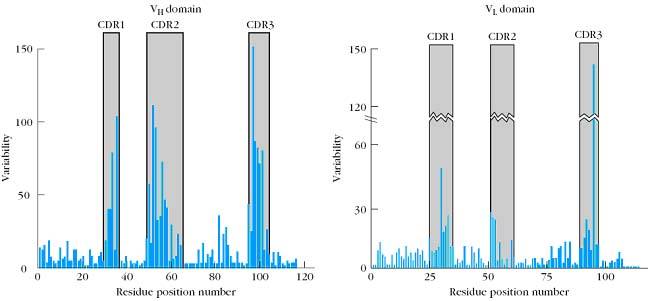
Detailed comparisons of the amino acid sequences of a large number of VL and VH domains revealed that the sequence variation is concentrated in a few discrete regions of these domains. The pattern of this variation is best summarized by a quantitative plot of the variability at each position of the polypeptide chain. The variability is defined as:
Number of different amino acids at a given position
Variability = ------------------------------------------------------------------------
Frequency of the most common amino acid at given position
Thus if a comparison of the sequences of 100 heavy chains revealed that a serine was found in position 7 in 51 of the sequences (frequency 0.51), it would be the most common amino acid. If examination of the other 49 sequences showed that position 7 was occupied by either glutamine, histidine, proline, or tryptophan, the variability at that position would be 9.8 (5/0.51).Variability plots of VL and VH domains of human antibodies show that maximum variation is seen in those portions of the sequence that correspond to the loops that join the b strands. These regions were originally called hypervariable regions in recognition of their high variability. Hyper variable regions form the antigen binding site of the antibody molecule. Because the antigen binding site is complementary to the structure of the epitope, these areas are now more widely called complementarity determining regions (CDRs). The three heavy-chain and three light-chain CDR regions are located on the loops that connect the b strands of the VH and VL domains. The remainder of the VL and VH domains exhibit far less variation; these stretches are called the framework regions (FRs). The wide range of specificities exhibited by antibodies is due to variations in the length and amino acid sequence of the six CDRs in each Fab fragment. The framework region acts as a scaffold that supports these six loops. The three-dimensional structure of the framework regions of virtually all antibodies analyzed to date can be superimposed on one another; in contrast, the hypervariable loops (i.e., the CDRs) have different orientations in different antibodies.
CDRs are determined by mainly three methods namely X-rat crystallographic method, affinity labeling and Kabat plot. The finding that CDRs are the antigen-binding regions of antibodies has been confirmed directly by high-resolution x-ray crystallography of antigen-antibody complexes. Crystallographic analysis has been completed for many Fab fragments of monoclonal antibodies complexed either with large globular protein antigens or with a number of smaller antigens including carbohydrates, nucleic acids, peptides, and small haptens. In addition, complete structures have been obtained for several intact monoclonal antibodies. X-ray diffraction analysis of antibody-antigen complexes has shown that several CDRs may make contact with the antigen, and a number of complexes have been observed in which all six CDRs contact the antigen. In general, more residues in the heavy-chain CDRs appear to contact antigen than in the light-chain CDRs. Thus the VH domain often contributes more to antigen binding than the VL domain.
The earliest evidence demonstrating the role of the hypervariable regions in binding antigen was obtained by L.Wofsy, H. Metzger and S.J. Singer using an experimental technique called affinity labeling. This technique used a chemically reactive hapten as a labeling reagent that could bind specifically to an antibody forming a covalent bond with neighboring residues within the antibody’s binding site. Identification of the amino acids covalently bound to the affinity label revealed that hypervariable amino acids within light and heavy chain CDRs constitute the antigen binding site. When the sequence of variable regions of both heavy and light chains are compared for many immunoglobins against different antigens and the variability plotted against amino acid positions, the plot is known as kabat plot. This plot clearly shows the presence of three HV regions and four frame work regions in both VL and VH domains.
HINGE REGION:
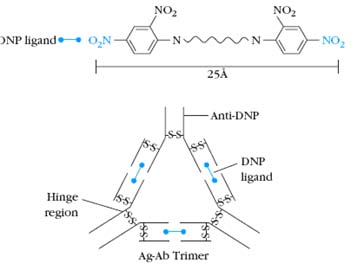
The g, d, and a heavy chains contain an extended peptide sequence between the CH1 and CH2 domains that has no homology with the other domains. This region, called the hinge region, is rich in proline residues and is flexible, giving IgG, IgD, and IgA segmental flexibility. As a result, the two Fab arms can assume various angles to each other when antigen is bound. This flexibility of the hinge region can be visualized in electron micrographs of antigen-antibody complexes. For example, when a molecule containing two dinitrophenol (DNP) groups reacts with anti-DNP antibody and the complex is captured on a grid, negatively stained, and observed by electron microscopy, large complexes (e.g., dimers, trimers, tetramers) are seen. The angle between the arms of the Y-shaped antibody molecules differs in the different complexes, reflecting the flexibility of the hinge region.
CLASSIFICATION:
Antibodies are classified mainly in two ways namely on the basis of constant region of heavy chain and antigenic determinants of antibody.
1. Classification on the Basis of Constant region of Heavy chain:
The antibody molecule is composed of 2 identical peptide chains. One pair, heavy chains, is at least twice as long as the other pair (light chains).The chains are held together by disulfide bonds and by ionic and hydrophobic forces. The heavy chains determine the class and are named with the Greek letter corresponding to the class. Subclasses of immunoglobulins can be distinguished on the basis of allotypes, which are single amino acid mutations in the constant region. There are 5 classes of immunoglobulins:
1. IgG-Model
2. IgM
3. IgA
4. IgD
5. IgE
General
Antibodies belong to the class of roughly spherical proteins called globulins. Since they are involved in immunity, they were given the class name immunoglobulin (abbreviated Ig). When immunologists wish to refer to antibodies in general regardless of their antigen specificity, they usually say immunoglobulins. When they speak of antibodies, they usually know the specific antigenic target involved.
Classes and Subclasses
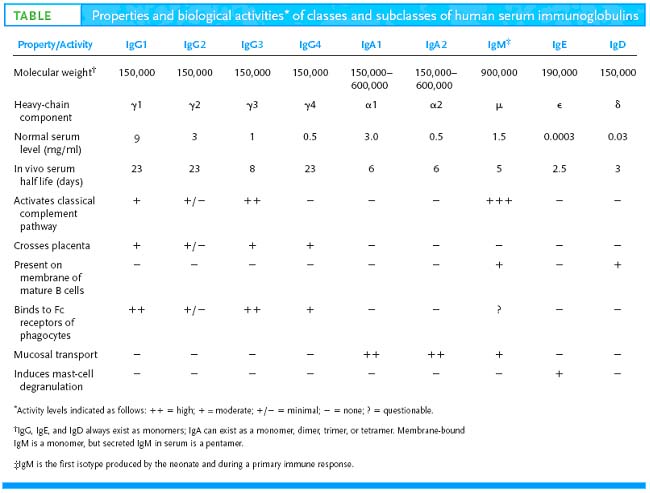
Igs can be subdivided on the basis of physical and functional properties into five classes, named for historical reasons IgG, IgM, IgA, IgD and IgE. Antibodies may act to eliminate antigens by five different mechanisms: neutralization, aggregation, opsonization, inducing complement mediated lysis, or inducing inflammation. Some classes are better at some mechanisms than other classes. As stated previously, there are five different Ig classes. IgG has four subclasses numbered 1 - 4; e.g., the third subclasses is IgG3. The different classes and subclasses are determined by differences between their heavy chains. As would be expected there are greater physical differences between the classes than between subclasses. The different classes and subclasses are called isotypes; i.e., protein families each member of which is represented in every member of the species. In practice, it is easier to distinguish between different IgG subclasses and between IgA subclasses by serological testing for markers unique to a given subclass. These markers arise due to single amino acid differences at particular sites in the constant region and are known as allotypic markers or allotypes. These markers are called Gm for IgG and Am for IgA. Even Igs of same type can be distinguished by directing antibodies against variable regions which are referred as idiotypes.
Immunoglobulin G – model
IgG was the first immunoglobulin to have its primary and secondary structures elucidated; therefore, its structure is considered the model for all immunoglobulins. (Porter and Edelman shared the Nobel Prize for this work.) The structures of other classes are described in terms of how they differ from or are similar to IgG.
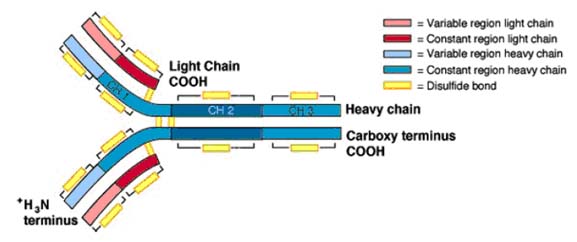
IgG is a bilaterally symmetrical tetramer composed of two identical heavy chains (molecular weight ~ 50,000 Daltons) and two identical light chains (MW ~ 25,000 Daltons). The chains are held together by disulfide bonds and by hydrophobic interactions. The number of disulfide bonds between the heavy chains and their exact location varies with the class and subclass of immunoglobulin. There is only one disulfide bond linking the heavy chain with the light chain which always involves the penultimate aminoacid on the light chain. The location of the linkage on the H chain also varies with the Ig class and subclass. For most classes and subclasses, it found around aminoacid 110 (of 440) of the heavy chain, but may be around amino acid 220. In humans, there are four subclasses of IgG numbered 1 - 4; e.g., Ig G1. The IgG heavy chains consist of about 440 amino acids exclusive of the area around the interchain disulfide bonds which contains a variable number depending on the subclass. Light chains also come in two physically distinct classes (or), but it has been more difficult to assign functional differences to the light chain classes. Both classes randomly associate with all classes and subclasses of immunoglobulins. Thus, a given IgG3 may have either or light chains but never both on the same molecule. The cysteine residue which forms the disulfide bond with the H chain is the next to the last (penultimate) amino acid in the L chain.
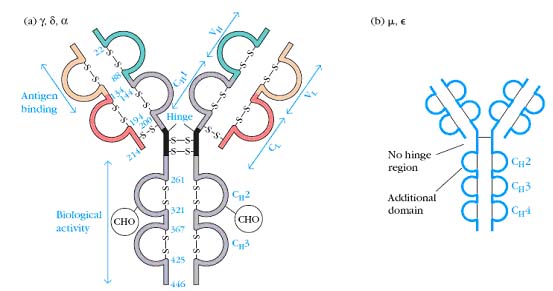
In 1965, Hilshmann, Craig and others determined the amino acid sequence of several L chains of the same class. When these sequences were compared, it was immediately apparent that the sequence of the carboxy terminal half approximately 110 amino acids was virtually identical; whereas, the amino terminal half was quite variable. These were named the constant or C region and the variable or V regions respectively. However, there were enough similarities between the two regions to establish that they must have evolved from the same original gene. For example, both regions have an intrachain disulfide bond at the same place within the region. As the primary sequence of heavy chains became available, a similar pattern emerged. The amino terminal 110 amino acids were relatively variable with the rest of the molecule consisting of three or four depending on the heavy chain class, repeats of approximately 110 amino acid sequences that have high (30+%) sequence homology within a subclass. The hinge region is the exception to this. These repeating units were very similar to that of the light chain constant regions, again suggesting that they evolved from the same primordial gene. Since the intrachain disulfide bonds hold each region in a somewhat spherical shape, the repeating units are called domains. Thus, IgG heavy chains have one variable domain (VH) and three constant domains (CH1, CH2, and CH3). Similarly, light chains have one variable domain (VL) and one constant domain (CL). This is seen in the three dimensional representation of the IgG molecule. Subsequent comparisons of many variable regions showed that most of the mutations were confined to three short stretches which were called hypervariable regions, one of which occurs where the variable and constant regions join. The antigen binding site is formed by a cleft at the end of the folded molecule where the heavy and light chain hypervariable regions come together.
The hinge region is the stretch between the CH1 and CH2 domains that contains the interchain disulfide bonds. Flexibility in this region allows the ends the molecule to move independently. The heavy chains are also glycosylated in this region which helps protect this somewhat exposed area against degradation. One of the real puzzles about Igs is how can the limited genetic material available code for proteins that have the ability to specifically recognize over ten million different antigens. This problem was solved only within the last few years. It turns out that antibodies are made by joining separate genes together. By randomly assorting the genes making up an antibody, the range of antigens that can be recognized is greatly increased.
Immunoglobulin M:
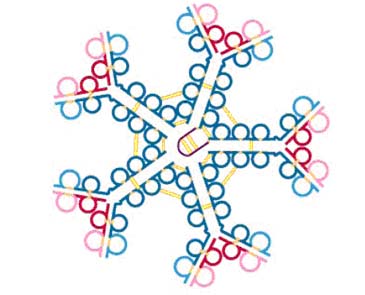
IgM in serum is composed of a pentamer of the basic 4 chain structure held together by inter H chain disulfide bonds. Each basic unit has the same amino acid sequence as the others in the pentamer. Thus, it has the ability to bind to 10 antigens at the same time but true valency is only five. In addition, its heavy chain ( ) is larger than the chain because it has four constant domains. Its molecular weight is about 65,000 Daltons. Thus, the molecular weight of the monomeric form of IgM is about 180,000 Daltons. The structure of IgM is similar to that of IgG except the IgM heavy chain has an extra domain. A small, cysteine-rich protein called J chain initiates cross linking of C3 and C4 of five IgM monomers to make the circulating, pentameric form of IgM. Polymerization is initiated by and the resulting pentamer is further stabilized by another molecule, the joining chain or J chain. J chain is a peptide of about 15,000 Daltons which is rich in cysteines that are available to form disulfide bonds. IgM is so large it is normally confined to the intravascular space. One of the benefits of the inflammatory response is it lets IgM escape into the tissue spaces at the site where it may be needed to bind potential pathogens.
Immunoglobulin A:
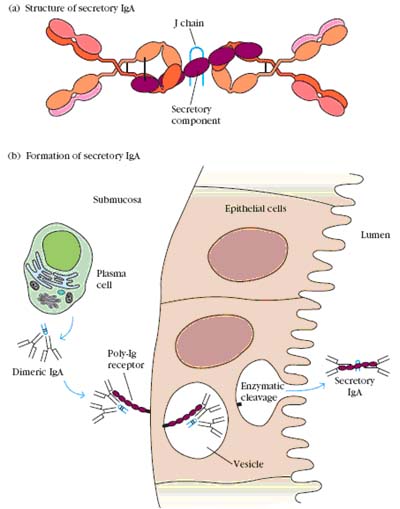
Serum IgA is similar in structure to IgG. Its heavy chain is a little larger, about 60,000 Daltons; although it contains only 3 constant region domains. It has two subclasses, IgA1 and IgA2. In blood and tissue fluids, IgA is a monomer, but in secretions, it is a dimer or sometimes higher polymer. As with IgM, polymerization is initiated and stabilized by J chain. IgA is actively secreted in tears, saliva, mucosal and gastric fluids, and milk. The transport protein which removes IgA from blood and carries it across epithelial cells to the secretory lumen is called secretory component. It is rich in cysteine residues which form numerous S-S bonds with the IgA heavy chains. Secretory component also seems to provide some protection against digestion in the GI tract.
Immunoglobulin D:
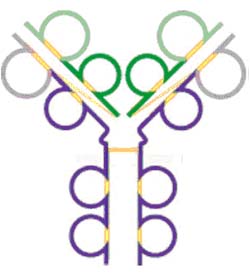
IgD consists of the basic 4 chain monomer similar to IgG except its heavy chain has a much longer hinge region resulting in a molecular weight of about 60,000 Daltons. Even with the longer hinge region, IgD seems to have only one inter-heavy chain disulfide bond. Because of this extended hinge region, it is very susceptible to being split apart by serum enzymes. IgD is similar to the structure of IgG.
Immunoglobulin E:
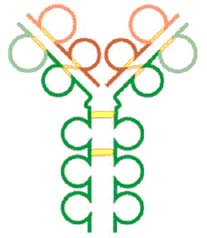
IgE also consists of the basic 4 chain unit. Its heavy chain is the largest of the immunoglobulins weighing 188,000 Daltons and having 4 C regions. The H chains are held together by two sets of intrachain disulfide bonds. It was the last to be discovered and characterized because it is present in such small amounts in serum (0.3 g/ml). IgE is similar to IgG except it has an extra constant region domain.
2. Classification based on antigenic determinants:
Since antibodies are glycoproteins, they can themselves function as potent immunogens to induce an antibody response. Such anti-Ig antibodies are powerful tools for the study of B-cell development and humoral immune responses. The antigenic determinants, or epitopes, on immunoglobulin molecules fall into three major categories: isotypic, allotypic, and idiotypic determinants, which are located in characteristic portions of the molecule.
Isotype:
Isotypic determinants are constant-region determinants that collectively define each heavy-chain class and subclass and each light-chain type and subtype within a species. Each isotype is encoded by a separate constant region gene, and all members of a species carry the same constant-region genes (which may include multiple alleles). Within a species, each normal individual will express all isotypes in the serum. Different species inherit different constant-region genes and therefore express different isotypes. Therefore, when an antibody from one species is injected into another species, the isotypic determinants will be recognized as foreign, inducing an antibody response to the isotypic determinants on the foreign antibody. Anti-isotype antibody is routinely used for research purposes to determine the class or subclass of serum antibody produced during an immune response or to characterize the class of membrane-bound antibody present on B cells.
Allotype:
Although all members of a species inherit the same set of isotype genes, multiple alleles exist for some of the genes. These alleles encode subtle amino acid differences, called allotypic determinants, that occur in some, but not all, members of a species. The sum of the individual allotypic determinants displayed by an antibody determines its allotype. In humans, allotypes have been characterized for all four IgG subclasses, for one IgA subclass, and for the light chain. The g-chain allotypes are referred to as Gm markers. Antibody to allotypic determinants can be produced by injecting antibodies from one member of a species into another member of the same species who carries different allotypic determinants.
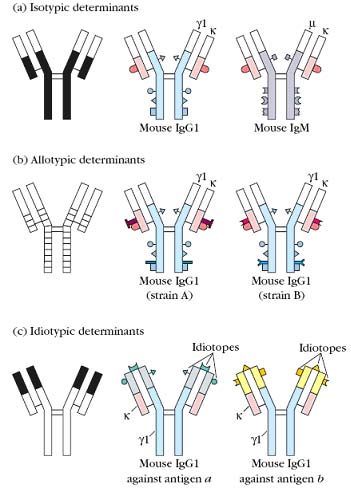
Idiotype:
The idiotypic determinants arise from the sequence of the heavy- and light-chain variable regions. Each individual antigenic determinant of the variable region is referred to as an idiotope. In some cases an idiotope may be the actual antigen-binding site, and in some cases an idiotope may comprise variable-region sequences outside of the antigen binding site. Each antibody will present multiple idiotopes; the sum of the individual idiotopes is called the idiotype of the antibody. Anti-idiotype antibody is produced by injecting antibodies that have minimal variation in their isotypes and allotypes, so that the idiotypic difference can be recognized.
MECHANISMS OF ACTION:
Immunoglobulin protective activities are expressed via several mechanisms.
a. Agglutination
b. Neutralization
c. Opsonization (IgG1 and 3)
d. Complement fixation (IgM and IgG)
e. Induction of inflammation (IgE)
Immunoglobulins perform their protective functions in five ways:
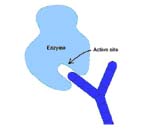
1. Direct action: They may bind to the active site of enzymes secreted by various bacteria which help them spread (e.g., collagenase) or to the binding site on viruses which would prevent them from attaching to their target.
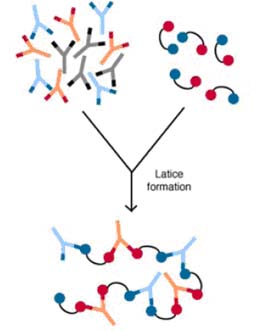
2. Agglutination or aggregation: Since all antibodies have at least two antigen binding sites, they can potentially bind to two organisms at the some time. Eventually, a large number of organisms could be clumped together in this fashion. These aggregated organisms would be less free to move, would have more trouble penetrating physical barriers and membranes, and usually would be engulfed more readily by macrophages which are not very effective at phagocytizing very small bacteria and all viruses. As would be expected, the polymeric immunoglobulins are more effective at agglutination than the others.
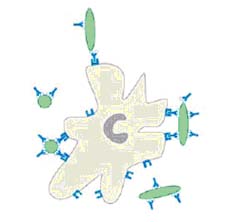
3. Opsonization: Macrophages and Neutrophils (PMNs) have surface receptors that recognize and bind to the Fc region of IgGl & IgG3. Therefore, anything coated with either of these antibodies will be bound by macrophages or PMNs and engulfed. One of the split products of complement, C3b, is also an opsonin, so IgM may indirectly contribute to opsonization via complement fixation.
4. Lysis: Complement activation can result in lysis of animal cells and gram negative organisms. Thus, complement fixing antibodies may indirectly cause lysis of susceptible targets.
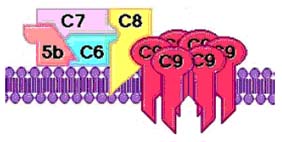
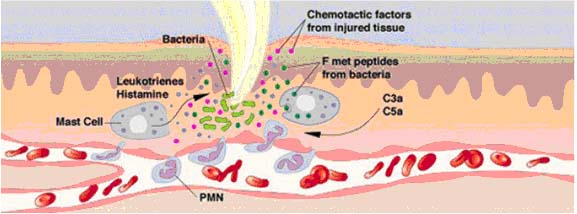
5. Inflammation: IgE stimulates an inflammatory response which is an important host defense mechanism. It is only with prolonged or extensive inflammation that the destructive effects outweigh the beneficial effects.
V. BIOLOGICAL FUNCTIONS OF IMMUNOGLOBULINS:
The biological functions of immunoglobulins vary with the class.
a. IgG is the most abundant immunoglobulin in blood and tissue fluids. It is an excellent opsonin and can activate complement.
b. IgM is the largest immunoglobulin which results in its being confined to intra vascular spaces. It appears first during phylogeny, ontogeny, and during an immune response. It is the most effective immunoglobulin in fixing complement.
c. IgA is actively secreted into external body fluids such as saliva where it acts to inhibit the entry of potential pathogens into the body.
d. IgD is part of the antigen-recognition/signaling mechanism on "virgin" B cells.
e. IgE is the immunoglobulin responsible for allergic reactions.
The basic function of immunoglobulins is to help the body protect itself against potential pathogens. The mechanisms by which they accomplish this vary with the Ig class and subclass as described previously.
Immunoglobulin G:
IgG is the most prevalent immunoglobulin in serum, contributing about 75% of the total circulating immunoglobulin. In adults it has a mean concentration of about 1250 mg/100 ml (mg %) with a normal range of 800-1700 mg%. It is also found in nearly equal concentrations in tissue fluids and lymph. IgGl and 3 are the immunoglobulins which are most responsible for labeling targets for removal by the phagocytic system (opsonins). Both macrophages and neutrophils have receptors for the Fc portion of both subclasses, so they are able to bind to and engulf organisms coated with these molecules. All subclasses except IgG4 can activate complement via the classical pathway, thereby causing the lysis of gram negative bacteria and animal cells. The immune system of newborn infants is still somewhat immature, so they are not immediately able to make IgG. Among other things, the ability of macrophages to process and present antigen is deficient. Babies start to make low levels of IgG by about three months and reaches 80% of adult levels in serum by age 12 months. All IgG subclasses except IgG2 can cross the placenta, so they provide the major protection against infection for the first 3 - 4 months of an infant's life. It is able to do this because it has a half-life in serum of ~ 23 days, which is the longest of any immunoglobulin. Declining maternal IgG levels in the newborn, and increasing amounts made by the baby cross around 3 - 4 months; i.e., the period of lowest IgG levels in the infant. This is the time during which the baby is most susceptible to low grade infections.
Immunoglobulin M:
IgM is the first immunoglobulin to appear during an immune response and is the first immunoglobulin one encounters as one ascends the phylogenetic tree. It is the most effective immunoglobulin at fixing complement and in aggregating targets. However, it does not serve as an opsonin. Because of its size, pentameric IgM is confined to the intact intravascular space but it is released during an inflammatory response. Its concentration in serum is about 125 mg% (range 65-120) for men and 160 mg% (80-320) for women. Its half-life is only about 5 days. Therefore, it is not nearly as important in host defense as IgG. However, monomeric IgM is the antigen receptor on B cells, thereby providing the specificity for the humoral immune response.
Immunoglobulin A:
IgA is actively secreted into various externalized body fluids; e.g., saliva, tears, gastric fluid, milk, and mucosal secretions. Because of this, it is present in secretions at higher concentrations than IgG. Since it is a dimer, it is very effective in aggregating targets which hampers their penetration of the body surface. In addition, aggregated IgA can activate complement via the alternative pathway. It is unclear whether this is an important protective function. Its concentration in serum is about 280 mg% (range 90- 450 mg %), and its half-life is about 6 days, so it can provide some internal protection as well.
Immunoglobulin E:
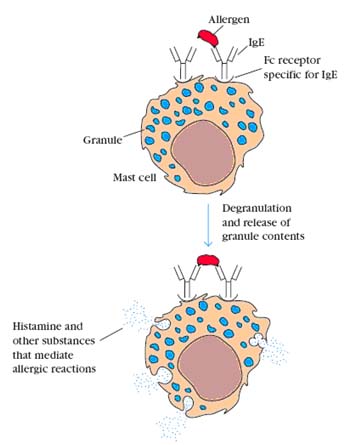
IgE is important in host defenses against parasitic infestations. It induces an inflammatory response which induces phagocytic cells to come into the area to deal with the potential pathogen. Unfortunately, it is also responsible for allergic responses. It is effective is extremely small amounts, being present in the serum at only 0.03 mg%. Mast cells have receptors for IgE, so they are coated with IgE from the plasma and tissue fluids. When these cell-bound IgE molecules bind to their target antigen (the allergen), they cause the mast cell to release a series of vasoactive compounds. The most important of these in humans is histamine which is why antihistamines are used in the treatment of hay fever and slow reactive substance of anaphylaxis which does a lot of the same things that histamine does but more slowly and persistently.
Immunoglobulin D:
It’s only known function is as part of the signaling complex of B cells. IgD is part of the antigen recognition unit of B cells. Its concentration about 3 mg%, is essentially unaffected by infections and it is probably not directly important in host defenses.
PROPERTIES OF ANTIBODIES: