DNA molecules, like all other biomolecules, can be damaged in numerous ways. Spontaneous damage due to replication errors, deamination, depurination and oxidation is compounded in the real world by the additional effects of radiation and environmental chemicals. The number of ways that DNA molecules can be damaged is very large. Since repair systems must be capable of recognizing and dealing with each type of damage, it is not surprising that there is a large number of different types of repair system.
Mutation, Mutagen, Mutagenesis and Mutant:
Any change in DNA base sequence referred as Mutation. Agent which causes mutation referred as Mutagen. The process through which mutation occurs known as Mutagenesis. Mutated genetic state of organisms are referred as Mutant. For example In E.coli Lac+ is wild type whereas Lac- is called as mutant.
TYPE OF MUTANTS:
1. Auxotrophic mutants
Mutation has inactivated a gene involved in the production of an essential metabolite, e.g. leu B6 E.Coli with this mutation require leucine to be supplemented to minimal media.
2. Conditional lethal mutants
Mutants that can only survive if cultured under a particular set of conditions, e.g. temperature sensitive . Mutations can only grow at permissive temperature (e.g. 30*C) and die at restrictive temperature (e.g. 376*C).
3. Antibiotic resistant mutants:
The target for the antibiotic becomes altered e.g. Streptomycin resistance due to a change in ribosomal protein S12.
4. Regulatory mutants
Have lost the ability to control expression. For example, constitutive mutants which express genes of the lac operon even in the absence of lactose e.g. mutation in the lacI repressor.
Mutagens:
A Spontaneous mutation occurs once in108 cells. The mutation rate can be increase by exposing cells to mutagens, which are either chemicals or physical agents such as UV-irradiation. Both chemicals and physical agents act by causing genetic damage that results in base changes in DNA. Depending upon the nature of mutagens they are classified into five types:
1. Base analogue
2. Chemical agents
3. Intercalating agents
4. Mutator genes
5. physical agents
1. Base analogue
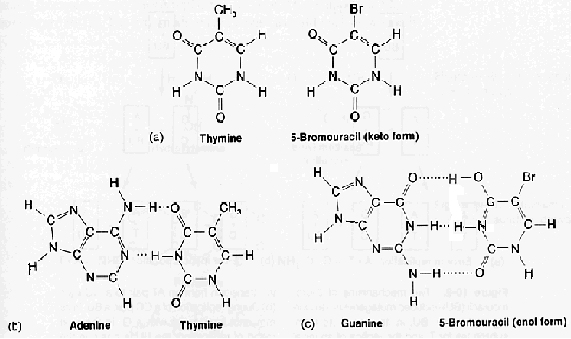
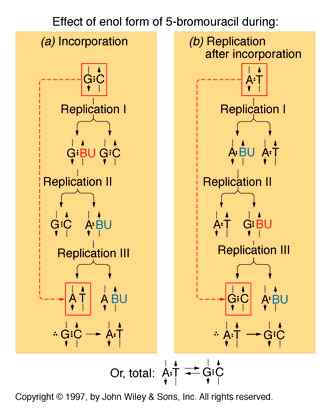
If a mutagen similar to one of the four bases required for the synthesis of DNA then the agents are known as base analogue. They substitutes for a standard base during replication and causes a new base pair to appear in daughter cells in a later generation. for example: 5- Bromo Uracil substituted instead of T or C which intern result in the conversion of A : T --> G:C and G:C --> A:T .

The base analogues used in Mutagenesis are substances that are sufficiently similar to naturally occurring DNA bases so that their deoxyribonucleotide triphosphates can be incorporated into DNA in place of the normal bases. However, they also have anomolous base-pairing properties, leading to an increased rate of mutagenesis. For example, 5-bromouracil pairs like thymine (5-methyluracil), but undergoes more enol tautomerization, leading to more frequent mispairing with guanine. Similarly, 2-aminopurine normally pairs with thymine, but can also pair with cytosine. These mispairing lead to an increase in the frequency of transitions.
2.Chemical mutagens:
A chemical mutagen is a substance that can alter a base that is already incorporated in DNA and thereby change its hydrogen-bonding specificity. Three commonly used chemical mutagens include nitrous acid, hydroxyl amine and ethyl methane sulfonate.
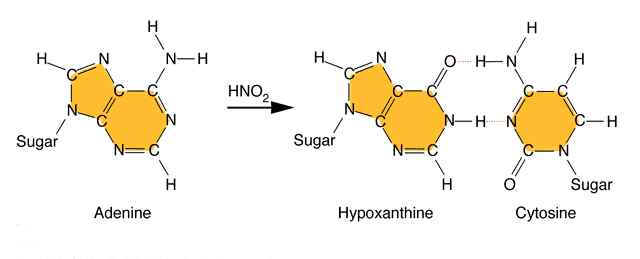
Nitrous acid: Treatment of DNA with nitrous acid leads to deamination of cytosine and adenine, again resulting in transitions.
Hydroxylating agents: Hydroxylamine adds a hydroxyl group to the amino group at position 4 of cytosine, causing it to pair with A instead of G. Hydroxylamine thus causes a very specific CG to TA transition.
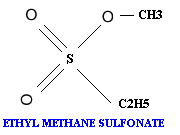
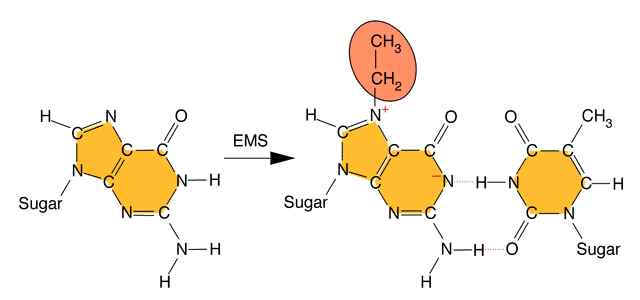
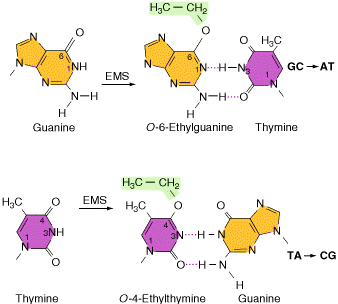
Alkylating agents: Certain alkylating agents, such as ethyl methane sulfonate (EMS) and ethyl ethane sulfonate (EES) add alkyl groups to purines, which can cause mispairing, and also destabilize the bond between the purine and deoxyribose, leaving apurinic sites. The absence of a base-pairing partner allows any base to be inserted during the next round of DNA synthesis. This frequently leads to transversions as well as transitions.
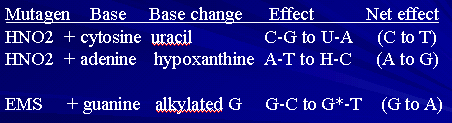
3. Intercalating agents:
These agents are planar ( flat) in structure and are approximately the same size as a purine-pyrimidine base pair. In solution they can insert between stacked base-pairs. The best known such agent is Ethidium bromide which is used to visualize DNA, as when it is inserted between the stacked base-pairs it fluoresces brightly to allow the DNA to be visualized. Replication of DNA containing intercalating agents is often seen to result in the addition of single bases, which seriously affects the reading frame of the gene. Proflavin and acridine orange are other intercalating agents. Their activity is because of planar ring structure.
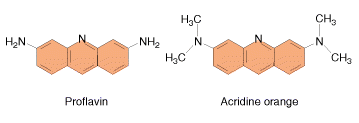
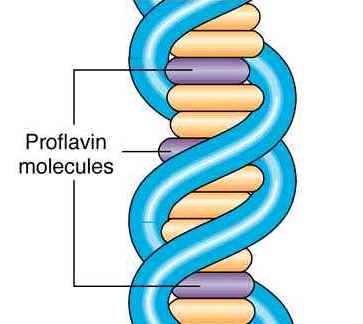
4. Mutator genes:
These are the certain genes which are usually plays vital role in the prevention of mutation. Defect in these genes might lead to the increased rate of mutation. Such genes are usually referred as mutator genes. Example for such genes include DNA methylase gene which actively involved in mismatch repair system.
5. physical agents:
Among the physical agents UV radiation and Ionizing radiation palys vital role in mutation.
i) UV radiations:
Ultraviolet light generates a number of photoproducts in DNA. Two different lesions that occur at adjacent pyrimidine residues include the cyclobutane pyrimidine photodimer (thymidine dimer) and the 6-4 photoproduct.
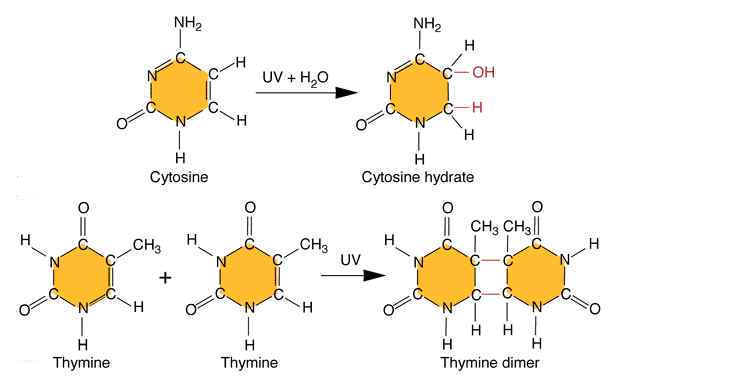
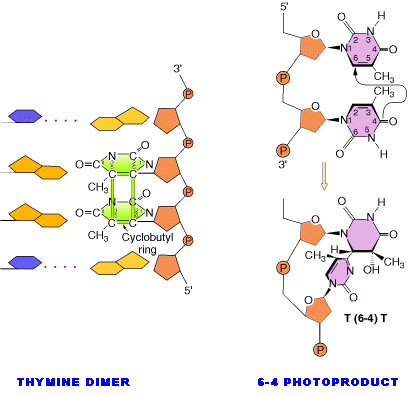
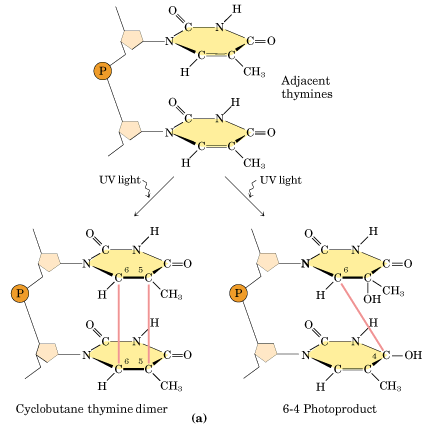
These lesions interfere with normal base pairing. These changes induces transitional, transversional and frameshift mutations indirectly.
Ultraviolet light is absorbed by the nucleic acid bases, and the resulting influx of energy can induce chemical changes. The most frequent photoproducts are the consequences of bond formation between adjacent pyrimidines within one strand, and, of these, the most frequent are cyclobutane pyrimidine dimers (CPDs). T-T CPDs are formed most readily, followed by T-C or C-T; C-C dimers are least abundant. One can obtain an idea of the extent of distortion of DNA chain structure caused by CPDs by noting that, in the diagram of a T-T CPD below, the cyclobutane ring, should have sides of approximately equal length. Thus the two adjacent pyrimidines must be pulled closer to each other than in normal DNA.
Dimers can also be produced by formation of a single covalent bond between the 6 position of one pyrimidine and the 4 position of the adjacent pyrimidine on the 3' side. The order of abundance of such pyrimidine (6-4) pyrimidine photoproducts (6-4PPs) is T-C>>C-C>T-T>C-T. Although only one bond attaches the adjacent pyrimidines, there is nevertheless extensive distortion of the normal DNA structure.
ii) Ionizing radiations:
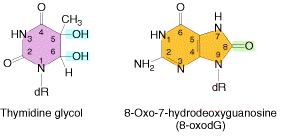
Ionizing radiation results in the formation of ionized and excited molecules that can cause damage to cellular components and to DNA. Because of the aqueous nature of biological systems, the molecules generated by the effects of ionizing radiation on water produce the most damage. Different types of oxygen radicals are produced. These include `OH, O2- and H2O2. These species can damage bases and cause different adducts and degradation products. Among these products, there are two products occurs mostly namely thymine glycol and 8-Oxodeoxy Guanosine.
TYPES OF MUTATION:
Mutation classified into different types depending upon several factors.
1. Depending upon the nature of occurrence, mutation are of two types namely Spontaneous mutation and Induced mutation. Spontaneous mutation occurs naturally without the involvement of any mutagen whereas if mutation occurs with the involvement of mutagen then they referred as Induced mutation.
1.1. SPONTANEOUS MUTATIONS:
Spontaneous mutations occurs mainly due to three reasons namely errors in replication, spontaneous lesions and transposable elements.
1.1.a. Errors in replication:
An error in DNA replication can occur when an illegitimate nucleotide pair forms during DNA replication. It intern leads to a base substitution. The main reason for the error in replication is due to the tautomeric shift of bases. Usually bases in DNA occur in many forms which differ in the positions of their atoms and in the bonds between the atoms. These form are in equilibrium. The keto form of each base is normally present in DNA, whereas the imino and enol forms of the bases are rare. Different forms of the bases listed below:
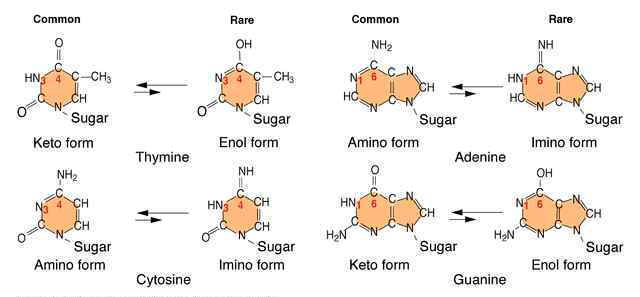
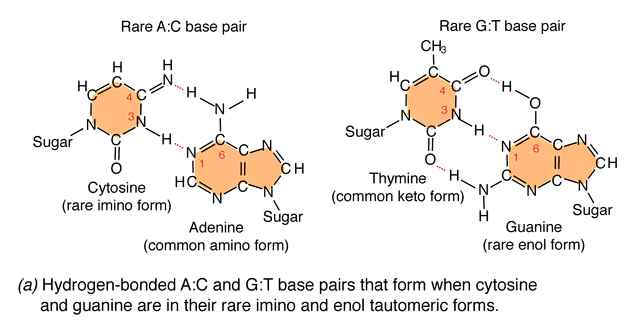
Due to the variation of bases from the usual form to rare form leads to mispairing in DNA. For example when cytosine changed into rare imino form then it is paired with Adenine. Similarly rare enol form of guanine makes it to pair with thymine. These type of changes referred as tautomeric shift. These type of changes intern leads to transitional, transversional and frameshift mutations in DNA.
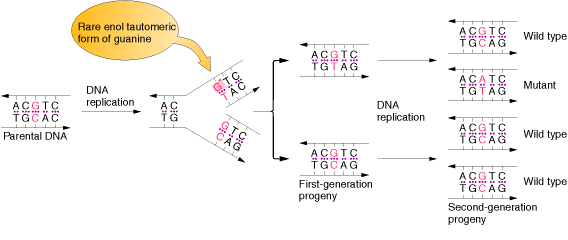
1.1.b. Spontaneous Lesions:
It is also another type of spontaneous mutation. It occurs mainly in two ways namely depurination and deamination.
1.1.b.1. Depurination:
It is the most common method of spontaneous Lesions. It occurs due to the breakage of glycosidic bond between the base and deoxyribose residues. Due to this, purine bases lost from the DNA. The frequency of depurination is one in 10,000 purines in DNA. Prolonged lesions might leads to severe genetic damages because the resulting apurinic sites cannot specify a base complementary to the original purine.
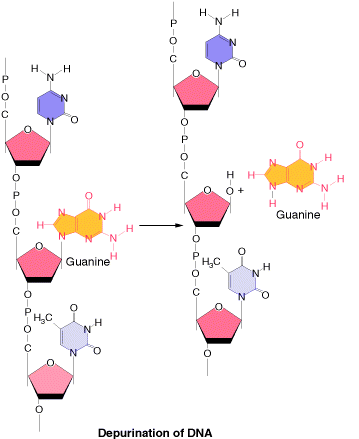
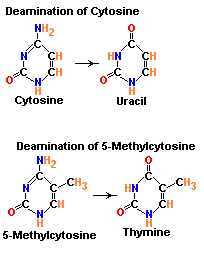
1.1.b.2. Demination:
Deamination refers to the removal of amino group from bases. Usually cytosine and 5-methyl cytosine undergoes deamination which result in the production of uracil and thymine respectively. Conversion of cytosine to uracil result in the transitional mutation GC --> AT similarly conversion of 5-methyl cytosine to thymine results in the transitional mutation GC --> AT.
1.1.c. Transposable Elements:
They are also otherwise known as "jumping genes" or Transposans. Due to the transfer of these components into the interior region of structural genes and regulatory genes leads to the abnormal expression of these genes. Thus these elements cause mutations.
1.2 INDUCED MUTATIONS:
This type of mutation is the major type. Mutation induced by the mutagen are found to be example for this type of mutation.
2. Depending upon the change in base sequence, mutation classified into different types namely, Point mutation, translocation, inversion and reversion.
2.1. Point mutation:
When only one base changes then the mutation referred as point mutation. This intern are of three types namely base substitution, base deletion and base insertion.
2.1.1. Base substitution:
Base substitution refers to the change of an single base in gene. It is of two types namely transition and transversion. When purine is replaced by another purine or pyrimidine is replaced by another pyrimidine then it is named as transition.
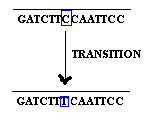
when purine replaced by an pyrimidine and vice versa then it is referred as transversion.
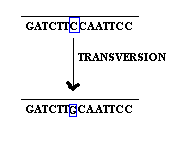
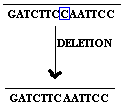
2.1.2. Base deletion:
Removal of Single base from the gene referred as base deletion.
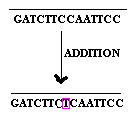
2.1.3. Base addition:
Addition of the single base to the gene referred as base addition.
2.2. other mutations:
Mutation can also occur with more than one base change like Deletion, Duplication, Inversion , Insertion and Translocation where fragment of DNA undergo changes.
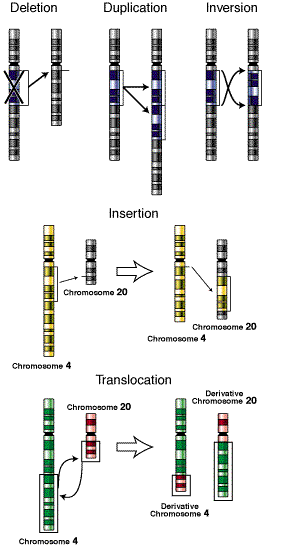
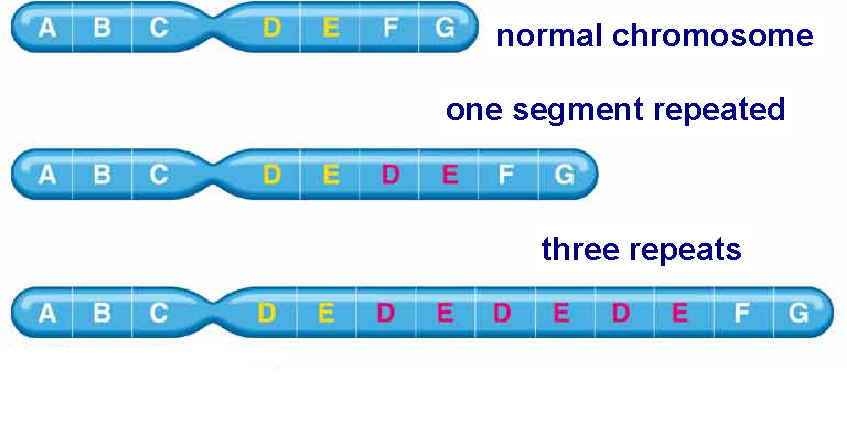
Deletion refers to the lose of fragment of DNA. Duplication refers to the doubling of fragment of DNA or repetition of specific DNA fragment two times. Inversion refers to change the polarity of fragment of DNA i.e. fragment of DNA removed and inverted then ligated to its original location. Insertion refers to the addition of fragment of DNA. Translocation refers to the transfer of fragment of DNA from its original location to new location.
2.3. Reversion:
Reversion refers to the type of mutation where the wild type of the genotype is regained due to mutation. It is otherwise known as back mutation or reverse mutation. If second mutation in a gene suppress the effect of first mutation then this type of mutations called as second site mutations or suppressor mutations. Reversion further divided into two types namely intergenic and intragenic reversion. The difference is mainly because of the position where second site mutation occurs. If it occurs in the same gene then it is called as intragenic reversion and if it occurs in other gene then it is called as intergenic reversion.
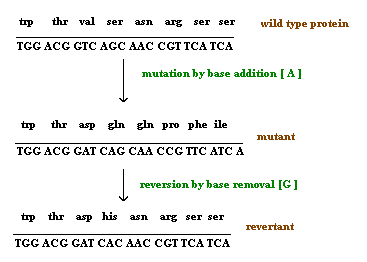
Depending upon the survival nature of the organism, mutation found to be of two types namely lethal and leaky mutations. If mutation prevent the growth of the organism then the mutation said to be of lethal mutation type and if mutation reduces the growth rate of the organisms then the muation said to be of leaky mutation type.
3. GENE MUTATION:
Gene mutation that results from changes within structure of a gene. It is sometimes called as a point mutation by considering that the change occurs at particular gene or point or loci in chromosome. Because of this mutation type, allele changes. Gene mutation broadly classified into two types depending upon the change with respect to wild type, namely forward mutation and reverse mutation.
when change converts wild type gene into mutant type then mutation referred as forward mutation whereas change converts mutant type into wild type then mutation referred as reverse or reversion or back mutation.
3.1. FORWARD MUTATION:
Forward mutation studied under two different headings namely
3.1.1. Single nucleotide pair (base pair) substitutions
3.1.2. Single nucleotide pair (base pair) additions and deletions
3.1.1. Single nucleotide pair (base pair) substitutions:
This mutation type studied under two different levels depending upon the molecule's nature namely at DNA level and Protein level. At DNA level, if purine or pyrimidine replaced by a different purine or pyrimidine respectively then the change referred as transition.
AT ------> GC GC ------> AT CG---------->TA TA------>CG
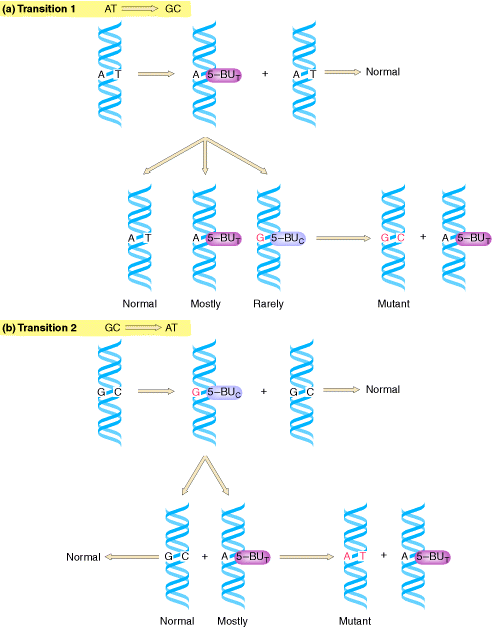
If purine replaced by another pyrimidine and vice versa then the change referred as transversion.
AT---------->CG AT---------->TA GC---------->TA GC ----------->CG
TA---------->GC TA---------->AT CG----------->AT CG---------->GC
At Protein level, depending upon the change occurring in the aminoacid nature in protein mutation studied under three different headings namely silent, missense and nonsense mutation.
If the mutation changes one codon for an aminoacid into another codon for that same amino acid then the mutation referred as silent mutation. In such mutation, there is no change occurs in structure and function of protein.
AGG ----------> CGG
This change of codon does not change amino acid because both codes for the same amino acid, arginine.
Due to mutation, if one amino acid is substituted by another amino acid then it is called as missense mutation. It occurs when codon for one amino acid is replaced by a codon for another amino acid. If a missense mutation causes the substitution of a chemically similar amino acid then the mutation is referred as synonymous substitution or mutation. This type of mutation results in a less severe effect on the protein's structure and function.
AAA---------->AGA
This results in a change of basic amino acid Lysine into another basic amino acid Arginine.
If a missense mutation causes the substitution of a chemically different amino acid then the mutation is called as non-synonymous substitution or mutation. It produce severe changes in protein structure and function. If such mutation occurs in amino acids contributing active site of protein then they are known as null (nothing) mutations.
AAA----------->GAA
This results in a change of basic amino acid Lysine into acidic amino acid Glutamic acid.
If codon for one amino acid is replaced by a translation termination codon or stop or nonsense codon then the mutation known as nonsense mutation. Non sense mutations will lead to the premature termination of translation. Thus, they have a considerable effect on protein function. Typically, unless it is very close to the 3' end of the open reading frame so that only a partly functional truncated polypeptide is produced, a nonsense mutation will produce a completely inactive protein product.
CAG ----------> UAG
This results in a change of amino acid, glutamine to a termination codon.
3.1.2. Single nucleotide pair (base pair) additions and deletions:
When a single nucleotide base is added or deleted, it would result in a change in amino acid sequence in protein after the site of mutation. This is because the sequence of mRNA is read by the translational apparatus in groups of three base pairs (codons). Hence these lesions are called frame shift mutations.
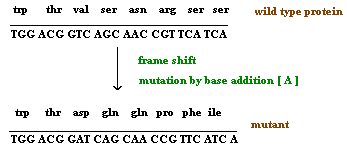
These mutations cause the entire aminoacid sequence translationally downstream of the mutant site to bear no relation to the original amino acid sequence. Thus, frameshift mutations typically exhibit complete loss of normal protein structure and function.
3.2. REVERSE MUTATION:
Reverse mutation classified into two types namely First site reversion and Second site reversion.
3.2.1. First site Reversion:
If reversion occurs at the same site i.e. in the same codon where previously mutation occurs, then it is referred as first site reversion. It of two types namely exact reversion and equivalent reversion. In exact reversion type, mutation result in the same site as it occurs in the wild type where as in equivalent reversion, it occurs in different site compared to the wild type but both mutation results in the formation of same amino acid as in the wild type or result in chemically similar aminoacid.
Exact reversion: AAA(Lys) ----------> GAA(Glu) ----------> AAA (Lys)
WILD TYPE MUTANT WILD TYPE
Equivalent reversion: UCC (Ser) ----------> UGC (Cys) ----------> AGC (Ser)
WILD TYPE MUTANT WILD TYPE
3.2.2. Second site reversion:
If reverse mutation occurs at the second site i.e. different codons, then such mutations referred as second site reversion or suppressor mutations. These suppressor mutations are further divided into two groups namely Intragenic and Extragenic suppressor mutations.
In Intragenic suppressor mutations, mutation occurs within the gene. The change occurring at the other codon suppresses the change produced by the previous mutation within the gene in this type of mutation.
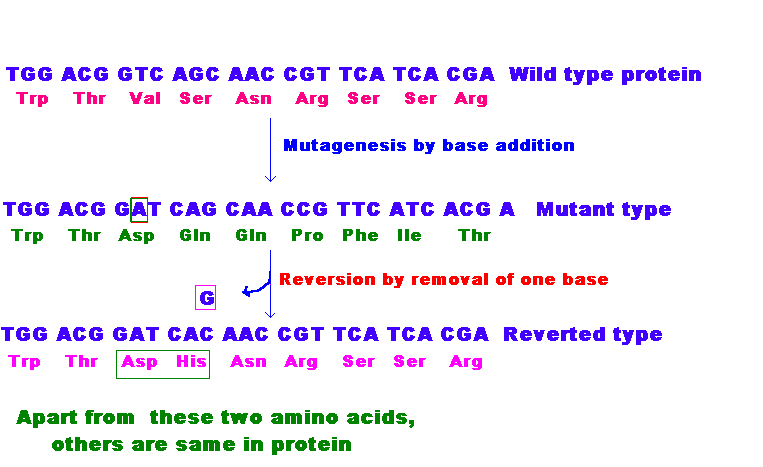
In Extragenic suppressor mutations, Second site mutation occurs at different genes. It is of four types namely Nonsense , Missense , Frameshift and Physiological suppressors.
If mutation in one gene makes other genes' stop codons to be considered as functional codons, then such mutations referred as Nonsense suppressor mutation. A gene (e.g. for tyrosine t-RNA) undergoes a mutational event in its anticodon region that enables the anticodon to recognize and align with a mutant nonsense codon (e.g. UAG) to insert an amino acid (e.g. tyrosine) and permit completion of the translation referred as Nonsense suppressor mutation.
If mutation in one gene changes the sequence of amino acids in another gene products then such type of mutations referred as Missense suppressors. It usually caused by change in tRNA anticodon. One missense suppressor in E.Coli is an abnormal tRNA that carries glycine but inserts it in response to arginine codons. Although wild type arginine codons are mistranslated, the observed mutations are not lethal, probably owing to the low efficiency of abnormal substitution.
If mutation in one gene cause the change in frame reading then such mutations referred as frameshift suppressor mutation. It occurs rarely. A four nucleotide anticodon in tRNA read a four letter codon which occurs by a single nucleotide pair insertion result in the change of amino acid sequence after the second site of mutation.
If mutation in one gene causes the change in the activity of cell carried out by another gene product then such mutation referred as Physiological suppressors. For example, an defect in one chemical pathway is circumvented by another mutation i.e. one that permits more efficient transport of a compound produced in smaller quantities owing to the original mutation.
Genes and chromosomes can mutate in either somatic or germinal tissue, and these changes are called somatic mutations and germinal mutations respectively. Somatic mutation in the Red Delicious apple and Germinal Mutation in an allele for curled ears are examples for somatic and germinal mutations respectively.
4. PHENOTYPIC CLASSIFICATIONS:
Depending upon the phenotypic consequences of mutations , it might be classified as Morphological, Lethal, Conditional, Biochemical , Loss of function and Gain of function Mutations.
Morphological Mutations:
When mutations affect the outwardly visible properties of an organisms then such mutations referred as Morphological mutations. For example: Curly wings in Drosophila and dwarf peas.
Lethal Mutations:
When mutations affect the survival of the organisms then such mutations are known as lethal mutations. For example: Mutation in all Hb gene
Conditional Mutations:
In the class of conditional mutations, a mutant allele causes a mutant phenotype in only a certain environment, called the restrictive condition, but causes a wild type phenotype in some different environment, called the permissive condition. Geneticists have studied many temperature conditional mutations. For example, certain Drosophila mutations are known as dominant heat sensitive lethal. Heterozygotes (H+/H) are wild type at 20*C (Permissive condition) but die if the temperature is raised to 30*C (the Restrictive condition).
Biochemical Mutations:
When mutations cause the loss or change of some biochemical function of the cell then such mutations are referred as Biochemical mutations. For example, one class of biochemically mutant fungi will not grow unless supplied with the nitrogenous base adenine. They are called ad mutations, whereas the wild type (Prototrophic) allele is ad+. Mutant ad alleles determine the auxotrophic, adenine-requiring phenotype.
Loss-of-function mutations:
When mutations result in the loss of function then such mutations known as Loss-of-function mutation. It is mostly found to be recessive in diploid cells. For example, mutation occur in alpha gene of Hb.
Gain-of-function mutations:
Sometimes mutations confers some new function on the gene then such mutations referred as gain-of-function.
5. CHROMOSOMAL MUTATIONS:
Any type of change in the chromosome structure or number is referred as chromosme aberration or chromosome mutation. Chromosome mutation is the process of change that results in rearranged chromosome parts, abnormal numbers of individual chromosomes, or abnormal numbers of chromosome sets. When chromosome mutation compared with gene mutation, later is applied both to the process and to the product. Sometimes chromosome mutations can be detected by microscopic examination, sometimes by genetic analysis and sometimes by both whereas gene mutations are never detectable microscopically on a chromosome. In general, chromosome mutation are of two types namely chromosomal structural mutations and chromosomal number mutations.
5.1. CHROMOSOMAL STRUCTURAL MUTATIONS:
Following are the few types of changes which result in chromosomal structural mutations.
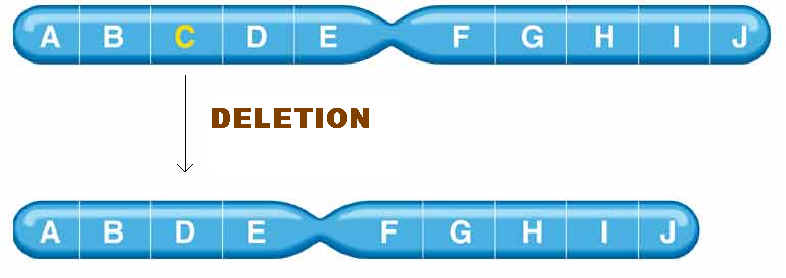
1.The simple loss of a chromosomal segment is called a deletion or deficiency. In the following diagram, region B has been deleted:
![]()
2. The presence of two copies of a chromosomal region is called a duplication:
![]()
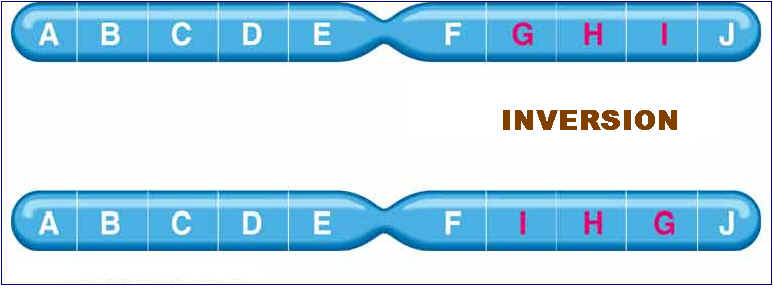
3. A segment of a chromosome can rotate 180 degrees and rejoin the chromosome, resulting in a chromosomal mutation called an inversion:
![]()
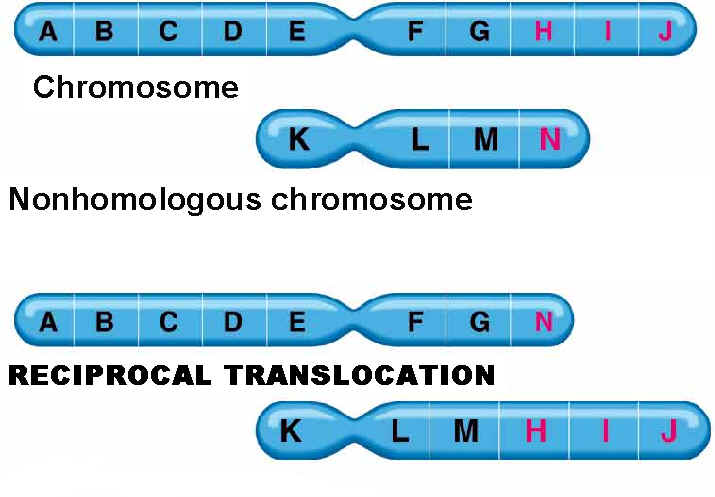
4. Finally, two nonhomologous chromosomes can exchange parts to produce a chromosomal mutation called a translocation:
![]()
MECHANISMS OF CHANGE:
These changes occurs mainly in two ways namely, by breakage and rejoining and by crossing over between repetitive DNA.
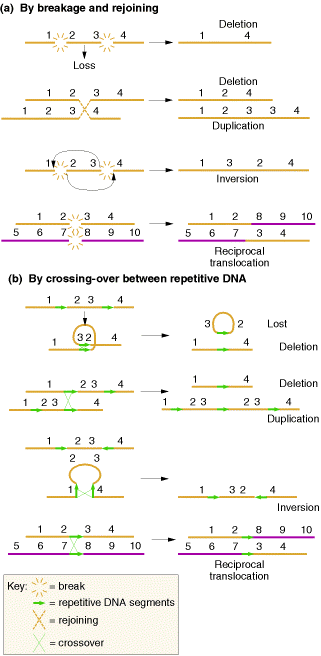
5.1.1. DELETIONS:
Deletions mainly are of four types namely interstitial deletion, terminal deletion, intragenic deletion and multigenic deletion. Break at two regions result in interstitial deletion; but breakage at terminal region result in terminal deletion. They are represented in the following figure.
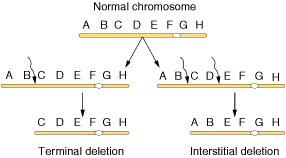
A small deletion within a gene, called as intragenic deletion which result in the inactivation of gene product. When deletions result in the loss of several genes then deletions referred as multigenic deletions.
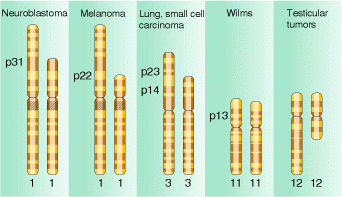
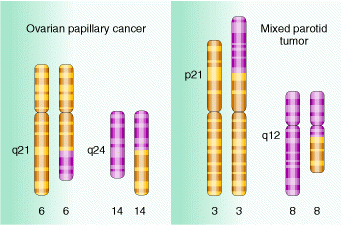
Deletions are recognized genetically by reduced recombinant frequency, Pseudodominance, recessive lethality and lack of reverse mutation and cytologically by deletion loops. Deletions result in several solid tumours which are indicated in the following figure:
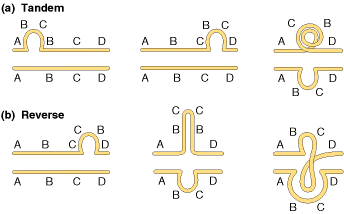
5.1.2. DUPLICATIONS:
Duplications are of two types i.e., tandem and reverse duplications. When duplication occur in a continuous region then it referred as tandem duplication where duplication occurs in reverse order then duplication referred as reverse duplication. Both of them explained in the following figure:
Duplications supply additional genetic material capable of evolving new functions.
5.1.3. INVERSIONS:
Inversion are of two types namely paracentric and pericentric inversions. If inversion occur outside the centromere then inversion referred as paracentric inversion whereas if inversion occur in the region spanning centromere them it is called as pericentric inversion. They are diagrammatically shown below:

Inversions determined during meiosis due to the formation of Inversion loop which is shown in the following figure:
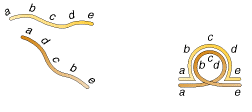
5.1.4. TRANSLOCATIONS:
Translocations leads to the transfer of fragments between two different chromosomes. Since parts are exchanged between two different chromosomes , it is otherwise known as reciprocal translocation. Translocations leads to the formation of different type of solid tumors in humans.
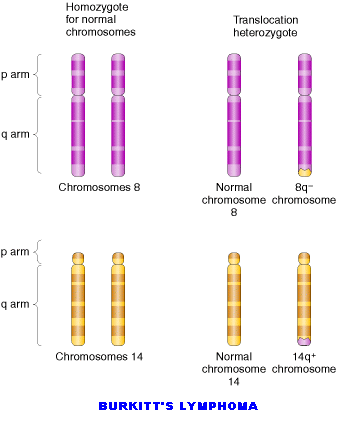
5.2. CHROMOSOMAL NUMBER MUTATIONS:
The number of chromosome is indicated by the suffix "Ploidy". There were different types of ploidies namely Euploidy, Monoploidy, Diploidy, Polyploidy and aneuploidy.
Euploid: When an organisms contains normal set of chromosomes then it is known as euploid.
Monoploid: When an organisms contain single set of chromosomes then it is referred as monoploid.
Diploid: When an organisms contain two set of chromosomes then it is referred as diploid.
Polyploid: When an organisms contain more than two set of chromosomes then it is called as Polyploid.
Aneuploid: When an organism contain chromosome number that differs from the normal chromosome number for the species by a small number of chromosomes then it is known as aneuploid.
5.2.1. MONOPLOIDY:
When organisms contain single set of chromosomes but the normal cell contain two set of chromosome then the condition referred as monoploidy. This is an chromosomal aberration because organism lacks single set of chromosome. Monoploid plants play an important role in modern approaches to plant breeding because Diploid nature is difficult to induce and select new gene mutations and study their effects. Monoploid plants are produced by tissue culture in the following steps: Pollen grains (haploid) are treated so that they will grow and are placed on agar plates containing certain plant hormones. Under these conditions, haploid embryoids will grow into monoploid plantlets. After having been moved to a medium containing different plant hormones, these plantlets will grow into mature monoploid plants with roots, stems, leaves, and flowers.
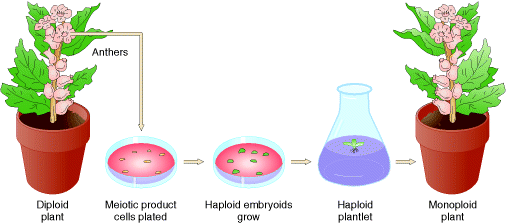
To produce new plant lines, geneticists produce monoploids with favorable genotypes and then double the chromosomes to form fertile, homozygous diploids.
5.2.2. POLYPLOIDS:
There are two types of polyploids namely autopolyploids, and allopolyploids. Autopolyploids composed of multiple sets of chromosomes from within one species. Allopolyploids composed of sets from different species which are closely related, however, the different chromosome sets are homeologous not fully homologous. Autotetraploids arise naturally by the spontaneous accidental doubling of a 2x genome to a 4x genome and Autotetraploids can be induced artificially through the use of colchicine. Autotetraploid plants are advantageous as commercial crops because in plants, the larger number of chromosome sets often leads to increased size. Cell size, fruit size, flower size and so forth , can be larger in the polyploid. Different ways of formation of tetraploids shown in the following figure:
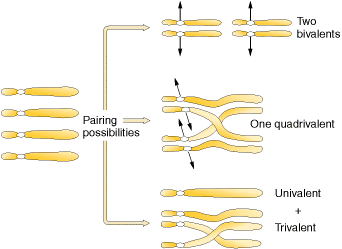
Of the three possibilities, two bivalents and one quadrivalent possibilities form tetraploids whereas univalent-trivalent combination yields non functional gametes.
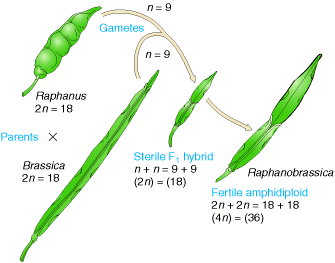
Allopolyploid synthesized to make a fertile hybrid varieties mainly. An allopolyploid fertile hybrid produced from the leaves of the cabbage and the roots of the radish. Each of these has 18 chromosomes and a viable hybrid progeny produced from seed. Most of the species found to be sterile whereas few species produce seeds. On planting, these seeds produced fertile individuals with 36 chromosomes. Allopolyploids possessing double chromosome number than their parental plant then the allopolyploids referred as amphidiploids.
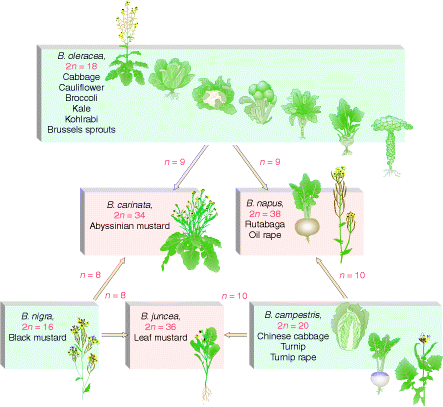
In nature, allopolyploidy seems to have been a major force in speciation of plants. In the following figure three different parent species have hybridized in all possible pair combinations to form new amphidiploid species.
Allopolyploids can be synthesized either by crossing related species and doubling the chromosomes of the hybrid or by asexually fusing the cells of different species.
5.2.3. ANEUPLOIDY:
Aneuploidy is the second major category of chromosome mutations in which chromosome number is abnormal. Aneuploid is an individual organism whose chromosome number differs from the wild type by part of a chromosome set. It has different conditions namely monosomic(2n-1), disoomic(n+1), trisomic(2n+1) and nullisomic(2n-2).
5.2.3.1. Monosomic:
It is the condition where one chromosome less than a normal chromosome count in a species. It is generally deleterious because of two reasons. First, If the lacking chromosome present in one copy then gene balance affected and Second, if a person is recessive to certain diseases then the complementary gene containing chromosome lost then the person become diseased. Nondisjuction in mitosis or meiosis is the cause of most aneuploids. Nondisjunction is a failure of chromosome segregation process, and two chromosomes go to one pole and none to the other.
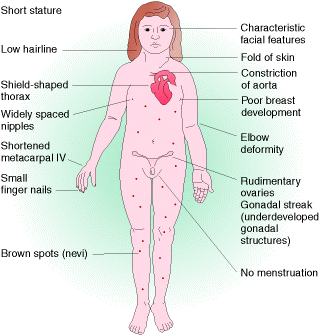
Turner Syndrome is an example for monosomic condition where one sex chromosome absent i.e. they have 44 autosomes + 1 X . Affected people have a characteristic, easily recognizable phenotype. They are sterile females, are short in stature, and often have a web of skin extending between the neck and shoulders. Although their intelligence is near normal, some of their specific cognitive functions are defective. About 1 in 5000 female births have this monosomic chromosomal complement. Monosomics for all human autosomes die in utero.
5.2.3.2. Disomic(n+1):
A disomic is an aberration of a haploid organism. In fungi, they can result from meiotic nondisjunction. Disomics in fungi can be selected from asci showing special spore abortion patterns or as meiotic progeny that must contain homologous chromosomes from both parents.
5.2.3.3. Trisomics(2n+1):
The trisomic condition also is one of chrosomal imbalance and can result in abnormality or death. However, there are many examples of viable trisomics. In other words trisomic individuals possesses one extra chromosome along with their two set of chromosomes.
Klinefelter syndrome is an example for trisomics in humans. These patients are males with lanky builds who are mentally retarded and sterile. Their chromosome make up is 44 autosomes + XXY.

Down Syndrome is also another trisomic disorder but in this case trisomy occur at the 21st chromosome set. It occuring at a frequency of about 0.15% of all live births.
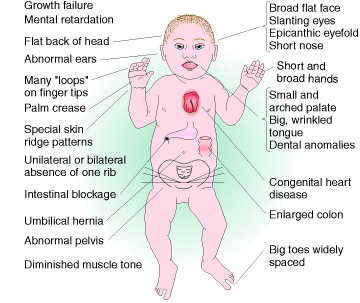
5.2.3.4. Nullisomics(2n-2):
It is an condition where two chromosome of a set in diploid found to be absent. This condition usually seen wheat plants. Although nullisomy is a lethal condition in diploids, an organism such as bread wheat, which behaves meiotically like a diploid although it is a hexaploid, can tolerate nullisomy.Their appearances differ from the normal hexaploids; furthermore, most of the nullisomics grow less vigorously.
REPAIR MECHANISMS:
When there is an damage occurs to DNA
then it should be repaired otherwise it will affect the present and future
generations. DNA, like any other molecule, can undergo a variety of
chemical reactions. Because DNA uniquely serves as a permanent copy of the cell
genome, however, changes in its structure are of much greater consequence than
are alterations in other cell components, such as RNAs or proteins. Mutations
can result from the incorporation of incorrect bases during DNA replication. In
addition, various chemical changes occur in DNA either spontaneously or as
a result of exposure to chemicals or radiation. Such damage to DNA can block
replication or transcription, and can result in a high frequency of mutations![]() consequences
that are unacceptable from the standpoint of cell reproduction. To maintain the
integrity of their genomes, cells have therefore had to evolve mechanisms to
repair damaged DNA. Repair system generally are of two types namely direct
reversal type , Excision repair , Recombinational repair and SOS Repair.
consequences
that are unacceptable from the standpoint of cell reproduction. To maintain the
integrity of their genomes, cells have therefore had to evolve mechanisms to
repair damaged DNA. Repair system generally are of two types namely direct
reversal type , Excision repair , Recombinational repair and SOS Repair.
Most damage to DNA is repaired by removal of the damaged bases followed by resynthesis of the excised region. Some lesions in DNA, however, can be repaired by direct reversal of the damage, which may be a more efficient way of dealing with specific types of DNA damage that occur frequently. Only a few types of DNA damage are repaired in this way, particularly pyrimidine dimers resulting from exposure to ultraviolet (UV) light and alkylated guanine residues that have been modified by the addition of methyl or ethyl groups at the O6 position of the purine ring.
1.1. PHOTO REACTIVATION:
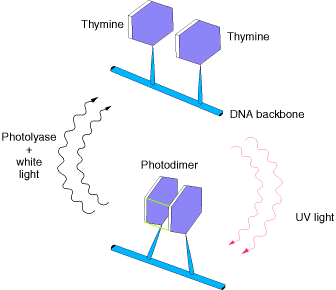
UV light is one of the major sources of damage to DNA and is also the most thoroughly studied form of DNA damage in terms of repair mechanisms. Its importance is illustrated by the fact that exposure to solar UV irradiation is the cause of almost all skin cancer in humans. The major type of damage induced by UV light is the formation of pyrimidine dimers, in which adjacent pyrimidines on the same strand of DNA are joined by the formation of a cyclobutane ring resulting from saturation of the double bonds between carbons 5 and 6 . The formation of such dimers distorts the structure of the DNA chain and blocks transcription or replication past the site of damage, so their repair is closely correlated with the ability of cells to survive UV irradiation. One mechanism of repairing UV-induced pyrimidine dimers is direct reversal of the dimerization reaction. The cyclobutane pyrimidine photodimer can be repaired by a photolyase that has been found in bacteria and lower eukaryotes but not in humans. The process is called photoreactivation because energy derived from visible light is utilized to break the cyclobutane ring structure . The original pyrimidine bases remain in DNA, now restored to their normal state. As might be expected from the fact that solar UV irradiation is a major source of DNA damage for diverse cell types, the repair of pyrimidine dimers by photoreactivation is common to a variety of prokaryotic and eukaryotic cells, including E. coli, yeasts, and some species of plants and animals. Curiously, however, photoreactivation is not universal; many species (including humans) lack this mechanism of DNA repair. This enzyme cannot operate in the dark, so other repair pathways are required to remove UV damage. A photolyase that reverses the 6-4 photoproducts has been detected in plants and Drosophila.
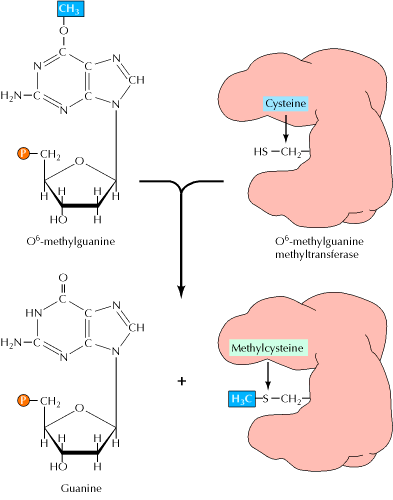
1.2. ALKYLTRANSFERASE REPAIR SYSTEM:
Another form of direct repair deals with damage resulting from the reaction between alkylating agents and DNA. Alkylating agents are reactive compounds that can transfer methyl or ethyl groups to a DNA base, thereby chemically modifying the base. A particularly important type of damage is methylation of the O6 position of guanine, because the product, O6-methylguanine, forms complementary base pairs with thymine instead of cytosine. This lesion can be repaired by an enzyme (called O6-methylguanine methyltransferase) that transfers the methyl group from O6-methylguanine to a cysteine residue in its active site. The potentially mutagenic chemical modification is thus removed, and the original guanine is restored. Enzymes that catalyze this direct repair reaction are widespread in both prokaryotes and eukaryotes, including humans.
2. EXCISION REPAIR SYSTEM:
Excision repair system includes methods
which repairs the damage by exchanging the damaged bases or
nucleotides. Although direct repair is an efficient way of dealing with
particular types of DNA damage, excision repair is a more general means of
repairing a wide variety of chemical alterations to DNA. Consequently, the
various types of excision repair are the most important DNA repair mechanisms in both
prokaryotic and eukaryotic cells. In excision repair, the damaged DNA is
recognized and removed, either as free bases or as nucleotides. The resulting
gap is then filled in by synthesis of a new DNA strand, using the undamaged
complementary strand as a template. Three types of excision repair![]() base-excision
repair, nucleotide-excision repair, and mismatch repair
base-excision
repair, nucleotide-excision repair, and mismatch repair![]() enable
cells to cope with a variety of different kinds of DNA damage.
enable
cells to cope with a variety of different kinds of DNA damage.
2.1. BASE EXCISION REPAIR SYSTEM:
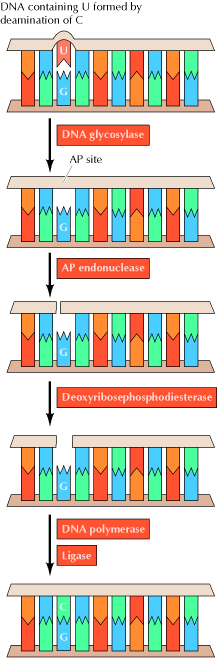
Uracil can arise in DNA by two mechanisms: (1) Uracil (as dUTP [deoxyuridine
triphosphate]) is occasionally incorporated in place of thymine during DNA
synthesis, and (2) uracil can be formed in DNA by the deamination of cytosine.
The second mechanism is of much greater biological significance because it
alters the normal pattern of complementary base pairing and thus represents a
mutagenic event. The excision of uracil in DNA is catalyzed by DNA
glycosylase, an enzyme that cleaves the bond linking the base (uracil) to
the deoxyribose of the DNA backbone. This reaction yields free uracil and an
apyrimidinic site![]() a
sugar with no base attached. DNA glycosylases also recognize and remove other
abnormal bases, including hypoxanthine formed by the deamination of adenine,
pyrimidine dimers, alkylated purines other than O6-alkylguanine, and
bases damaged by oxidation or ionizing radiation.
a
sugar with no base attached. DNA glycosylases also recognize and remove other
abnormal bases, including hypoxanthine formed by the deamination of adenine,
pyrimidine dimers, alkylated purines other than O6-alkylguanine, and
bases damaged by oxidation or ionizing radiation.
The result of DNA glycosylase action is the formation of an apyridiminic or apurinic site (generally called an AP site) in DNA. Similar AP sites are formed as the result of the spontaneous loss of purine bases, which occurs at a significant rate under normal cellular conditions. For example, each cell in the human body is estimated to lose several thousand purine bases daily. These sites are repaired by AP endonuclease, which cleaves adjacent to the AP site. The remaining deoxyribose moiety is then removed, and the resulting single-base gap is filled by DNA polymerase and ligase.
2.1.1. GO REPAIR SYSTEM:
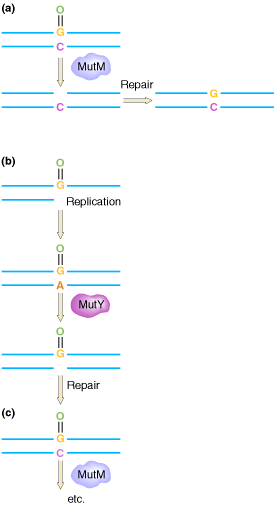
The GO REPAIR system acts in three different ways. They are as follows:
(a) 8-OxodG lesions are removed by the MutM protein, leaving an AP site that is repaired by endonucleases and repair synthesis.
(b) However, when replicating polymerases are allowed to
operate across from the lesion, they usually add an A residue. This mispair
would result in the GC ![]() TA change, but the MutY protein removes the A, allowing repair of the resulting
AP site.
TA change, but the MutY protein removes the A, allowing repair of the resulting
AP site.
(c) When repair polymerases operate across from the 8-oxodG lesion, they preferentially restore a C across from the lesion, allowing the MutM protein another opportunity to remove the lesion.
The mutT product prevents incorporation of GO across from A. The human counterparts of the mutT, mutY, and mutM gene products have been detected.
2.2. NUCLEOTIDE EXCISION REPAIR SYSTEM:
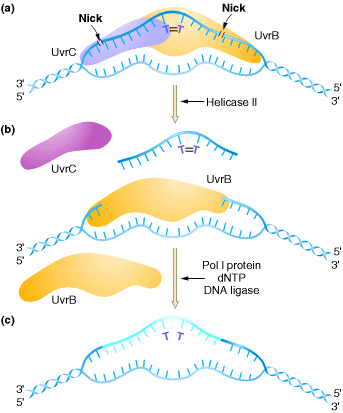
2.2.1. UvrABC Excinuclease system:
In E. coli, nucleotide-excision repair is
catalyzed by the products of three genes (uvrA, B, and C)
that were identified because mutations at these loci result in extreme
sensitivity to UV light. The protein UvrA recognizes damaged DNA and recruits
UvrB and UvrC to the site of the lesion. UvrB and UvrC then cleave on the 3![]() and 5
and 5![]() sides of the damaged site, respectively, thus excising an oligonucleotide
consisting of 12 or 13 bases. The UvrABC complex is frequently called an excinuclease,
a name that reflects its ability to directly excise an oligonucleotide.
The action of a helicase is then required to remove the damage-containing
oligonucleotide from the double-stranded DNA molecule, and the resulting gap is
filled by DNA polymerase I and sealed by ligase.
sides of the damaged site, respectively, thus excising an oligonucleotide
consisting of 12 or 13 bases. The UvrABC complex is frequently called an excinuclease,
a name that reflects its ability to directly excise an oligonucleotide.
The action of a helicase is then required to remove the damage-containing
oligonucleotide from the double-stranded DNA molecule, and the resulting gap is
filled by DNA polymerase I and sealed by ligase.
The human excinuclease is considerably more complex than its bacterial counterpart and includes at least 17 proteins. However, the basic steps are the same as those in E. coli.
2.2.2. EXCISION REPAIR SYSTEM IN EUKARYOTES:
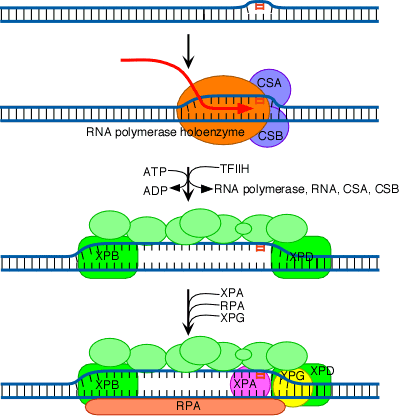
In mammalian cells, the XPA protein (and possibly also XPC)
initiates repair by recognizing damaged DNA and forming complexes with other
proteins involved in the repair process. These include the XPB and XPD proteins,
which act as helicases that unwind the damaged DNA. In addition, the binding of
XPA to damaged DNA leads to the recruitment of XPF (as a heterodimer with ERCC1)
and XPG to the repair complex. XPF/ERCC1 and XPG are endonucleases, which cleave
DNA on the 5![]() and 3
and 3![]() sides of the damaged site, respectively. This cleavage excises an
oligonucleotide consisting of approximately 30 bases. The resulting gap then
appears to be filled in by DNA polymerase d or
e (in association with RFC and PCNA) and
sealed by ligase.
sides of the damaged site, respectively. This cleavage excises an
oligonucleotide consisting of approximately 30 bases. The resulting gap then
appears to be filled in by DNA polymerase d or
e (in association with RFC and PCNA) and
sealed by ligase.
The initial steps depend on whether the damage is in the actively transcribed strand of a gene or elsewhere in the genome. If the damage is not in the actively transcribed strand of a gene, then the damage is recognized and bound by a heterodimer consisting of the XPC and HR23B proteins. The binding of XPC and HR23B initiates the process of "global genome repair" (GGR), which simply means repair anywhere in the genome.
The XPC/HR23B dimer appears to recognize damaged DNA based on the extent of distortion of the normal helical DNA structure caused by the damage. In the process of binding to the damaged region, XPC/HR23B is thought to further increase the extent of structural distortion, as illustrated in this diagram:
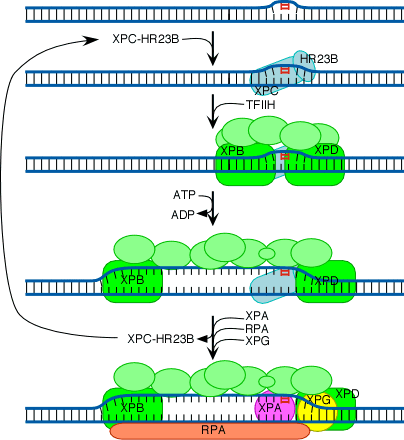
The increased distortion produced by XPC/HR23B permits the entry and binding of the general transcription factor TFIIH, whose 10 subunits are colored in various shades of green in the above diagram. Two of these subunits (XPB and XPD) are helicases, which bind to the damaged strand and use the energy of ATP to unwind a stretch of 20-30 nucleotides including the damaged site.
Three additional proteins then bind to and stabilize the open complex. The precise role of XPA is unclear, but evidence suggests that it checks to confirm that damage is present in the opened region and assists in stabilizing the open complex. RPA is the major eukaryotic single-stranded-DNA-binding protein. It binds to and protects both of the separated strands in the open complex. For clarity in the diagram, it is shown binding only to the bottom strand. XPG is a structure-specific nuclease.
Concomitant with the binding of XPA, RPA and XPG, XPC and HR23B are released. These two proteins are then free to recycle to other damaged sites where the repair process has not yet been initiated.
The next step in the repair process, for both GGR and TC-NER, is recruitment of another structure-specific endonuclease, the XPF-ERCC1 heterodimer:
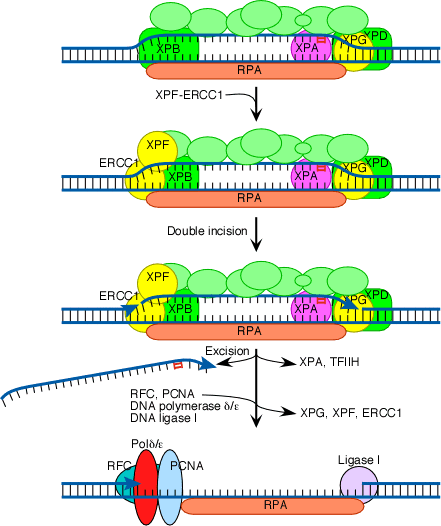
Both XPG and XPF-ERCC1 are specific for junctions between single- and double-stranded DNA. XPG, which is closely related to the FEN-1 nuclease that participates in base excision repair, cuts on the 3' side of such a junction, while ERCC1/XPF (a heterodimeric protein complex) cuts on the 5' side. The cut made by XPG is 2-8 nucleotides from the lesion, and the cut made by ERCC1/XPF is 15-24 nucleotides away. These distances are paired with each other (probably as a consequence of the structure of the multiprotein complex) in such a way that the damage-containing oligonucleotide between the cuts averages 27 nucleotides (range 24-32 nucleotides).
The damage-containing oligonucleotide is displaced concomitant with the binding of replicative gap-repair proteins (RFC, PCNA, DNA polymerase delta or epsilon), with the displacement of TFIIH, XPA, XPG, and XPF-ERCC1, and with new DNA synthesis that fills the gap. The final nick is sealed by DNA ligase I.
2.2.3. REPAIR COUPLING WITH TRANSCRIPTION:
An intriguing feature of nucleotide-excision repair is its relationship to transcription. A connection between transcription and repair was first suggested by experiments showing that transcribed strands of DNA are repaired more rapidly than nontranscribed strands in both E. coli and mammalian cells. Since DNA damage blocks transcription, this transcription-repair coupling is thought to be advantageous by allowing the cell to preferentially repair damage to actively expressed genes.
2.2.3.1. COUPLING IN PROKARYOTES:
In E. coli, the mechanism of transcription-repair coupling involves recognition of RNA polymerase stalled at a lesion in the DNA strand being transcribed. The stalled RNA polymerase is recognized by a protein called transcription-repair coupling factor, which displaces RNA polymerase and recruits the UvrABC excinuclease to the site of damage which intern removes the error and the transcription continued.
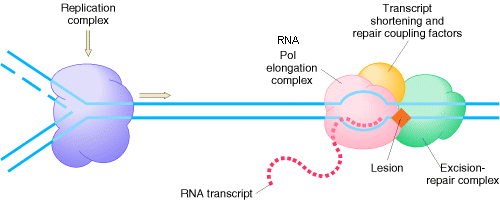
2.2.3.2. COUPLING IN EUKARYOTES:
In Cockayne's syndrome , two proteins found to be inactive namely CSA and CSB and they are also unable to carry out transcription blocked DNA repair. When transcription stahled at the thymidine dimer damaged site, CSA and CSB bound to stahled RNA polymerase. This then recruits the binding of TFII H factor to the site but RNA , RNA polymerase, CSA and CSB are released. The remaining reactions are same as in Nucleotide Excision repair system of eukaryotes. Thus CSA and CSB similar to XP C and HR23B in function.
2.3. MISMATCH REPAIR SYSTEM:
Some repair pathways are capable of recognizing errors even after DNA replication has already occurred. One such system, termed the mismatch repair system, can detect mismatches that occur in DNA replication. It occurs mainly in three steps namely
1. Recognize mismatched base pairs.
2. Determine which base in the mismatch is the
incorrect one.
3. Excise the incorrect base and carry out repair
synthesis.
The second point is the crucial property of such a system.
Unless it is capable of discriminating between the correct and the incorrect
bases, the mismatch repair system
could not determine which base to excise. If, for example, a G![]() T
mismatch occurs as a replication error, how can the system determine whether G
or T is incorrect? Both are normal bases in DNA. But replication errors produce
mismatches on the newly synthesized strand, so it is the base on this strand
that must be recognized and excised.
T
mismatch occurs as a replication error, how can the system determine whether G
or T is incorrect? Both are normal bases in DNA. But replication errors produce
mismatches on the newly synthesized strand, so it is the base on this strand
that must be recognized and excised.
To distinguish the old, template strand from the newly synthesized strand, the mismatch repair system in bacteria takes advantage of the normal delay in the postreplication methylation of the sequence
The methylating enzyme is adenine methylase, which creates 6-methyladenine on each strand. However, it takes the adenine methylase several minutes to recognize and modify the newly synthesized GATC stretches. During that interval, the mismatch repair system can operate because it can now distinguish the old strand from the new one by the methylation pattern. Methylating the 6-position of adenine does not affect base pairing, and it provides a convenient tag that can be detected by other enzyme systems. The old strand is methylated at GATC sequences right after replication. When the mismatched site has been identified, the mismatch repair system corrects the error.
2.3.1. MISMATCH REPAIR IN E.COLI:
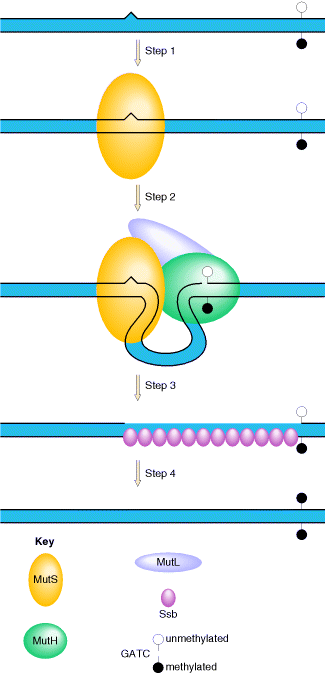
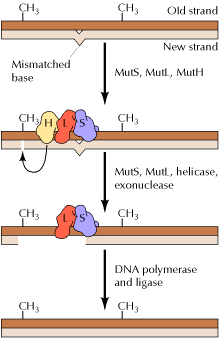
In E. coli, the ability of the mismatch repair system to distinguish between parental DNA and newly synthesized DNA is based on the fact that DNA of this bacterium is modified by the methylation of adenine residues within the sequence GATC to form 6-methyladenine. Since methylation occurs after replication, newly synthesized DNA strands are not methylated and thus can be specifically recognized by the mismatch repair enzymes. Mismatch repair is initiated by the protein MutS, which recognizes the mismatch and forms a complex with two other proteins called MutL and MutH. The MutH endonuclease then cleaves the unmethylated DNA strand at a GATC sequence. MutL and MutS then act together with an exonuclease and a helicase to excise the DNA between the strand break and the mismatch, with the resulting gap being filled by DNA polymerase and ligase.
2.3.2. MISMATCH REPAIR SYSTEM IN HUMANS:
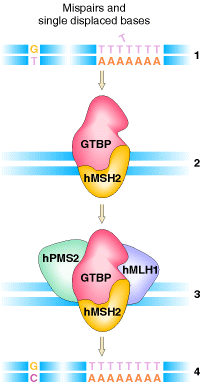
The mismatch repair
system has also been characterized in humans. Two of the proteins, hMSH2 and
hMLH1, are very similar to their bacterial counterparts, MutS and MutL,
respectively. The hMSH2 protein, together with the G![]() T-binding
protein (GTBP), binds to the mismatches and then recruits the other components
of the system, hPMS2 and hMLH1, to effect repair
of the mismatch.
T-binding
protein (GTBP), binds to the mismatches and then recruits the other components
of the system, hPMS2 and hMLH1, to effect repair
of the mismatch.
3. RECOMBINATION REPAIR:
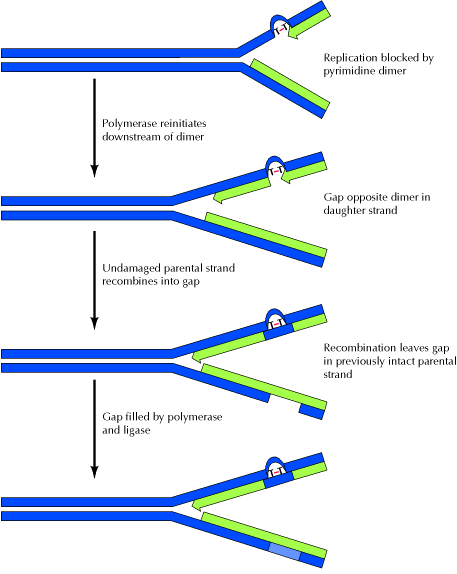
The presence of a thymine dimer blocks replication, but DNA polymerase can bypass the lesion and reinitiate replication at a new site downstream of the dimer which is referred as postdimer initiation. The result is a gap opposite the dimer in the newly synthesized DNA strand. In recombinational repair, this gap is filled by recombination with the undamaged parental strand. Although this leaves a gap in the previously intact parental strand, the gap can be filled by the actions of polymerase and ligase, using the intact daughter strand as a template. Two intact DNA molecules are thus formed, and the remaining thymine dimer eventually can be removed by excision repair. It is otherwise known as daughter strand gap or sister strand gap repair system because only the gaps formed opposite to dimers, rather than the dimers themselves, are repaired. Since recombination repair occurs after DNA replication, in contrast with excision repair, it has been called as postreplicational repair.
4. SOS REPAIR:
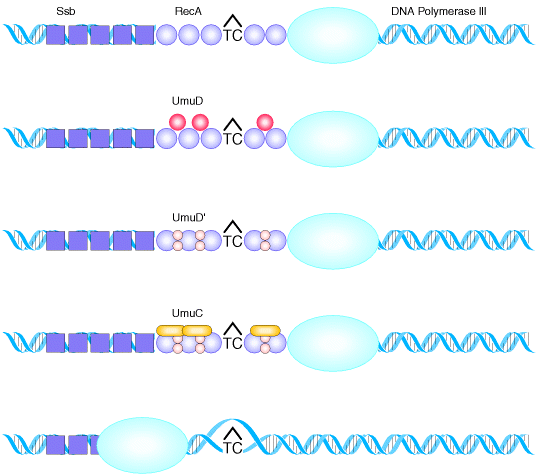
This is an example for the repair
system which found to be an error prone repair system. It is an error
prone process because the strands contain incorrect base eventhough intact DNA
strands are formed. This repair system otherwise known as By pass repair
system because it allows DNA chain growth across damaged segments at the cost of
fidelity of replication. The name SOS means Save Our Soul. This is
because, the principle involved is that survival with mutations is better than
no survival at all. DNA polymerase III, stops at a noncoding lesion, such as the T![]() C
photodimer, generating single-stranded regions that attract the Ssb
protein and RecA which forms filaments. The presence of RecA filaments
helps to signal the cell to synthesize UmuD, which is cleaved by RecA to yield
UmuD
C
photodimer, generating single-stranded regions that attract the Ssb
protein and RecA which forms filaments. The presence of RecA filaments
helps to signal the cell to synthesize UmuD, which is cleaved by RecA to yield
UmuD![]() .
The UmuC is recruited to form a complex with UmuD
.
The UmuC is recruited to form a complex with UmuD![]() that permits DNA polymerization to proceed past the blocking lesion. Since
DNA synthesis occur even across the thymine dimer region (damaged region),
synthesis of DNA referred as transdimer synthesis.
that permits DNA polymerization to proceed past the blocking lesion. Since
DNA synthesis occur even across the thymine dimer region (damaged region),
synthesis of DNA referred as transdimer synthesis.
The survival of an ultraviolet-irradiated lamda virus is higher on an irradiated host than on an unirradiated host when the damage is repaired by SOS repair. Then the phenomenon, is called as UV-reactivation or W reactivation. This is possible only in rec+ species only.
PREVENTION OF DNA ERROR:
Some enzymatic systems neutralize potentially damaging compounds before they even react with DNA. One example of such a system is the detoxification of superoxide radicals produced during oxidative damage to DNA: the enzyme superoxide dismutase catalyzes the conversion of the superoxide radicals into hydrogen peroxide, and the enzyme catalase, in turn, converts the hydrogen peroxide into water. Another error-prevention pathway depends on the protein product of the mutT gene: this enzyme prevents the incorporation of 8-oxodG, which arises by oxidation of dGTP, into DNA by hydrolyzing the triphosphate of 8-oxodG back to the monophosphate.
AMES TEST:
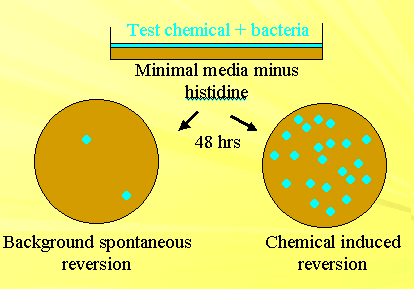
This is an important test which is usually used to test the mutagenicity of chemical substances. It is called so because of the formator of this test. It is based on histidine-requiring mutants (his- ) of Salmonella typhimurium.
In this test, first chemical to be tested mixed with the his mutant and cultured in a medium of Minimal media without histidine. Along with this, an control also setup where mutant alone grown in Minimal media without histidine. After 48 hours colonies grown were counted . Depending upon the mutagenicity nature of the chemical number of colonies varied. Increased colonies refers to the high mutagenic nature whereas decreased number indicated low mutagenic nature of the chemical.
The test measures mutagenicity by an increase in the frequency of spontaneous reversion of his- and his+ . This test is used to test industrial chemicals, food additives, pesticides, hair dyes and cosmetics. This test reduces the need of animal testing.